A Flexible Route to Synthesis and Molecular Docking of Some New Quinoline Derivatives through Imine and Cyclization Processes
Layla A. Othman1* , Shireen R. Mohammed2 and Maher K. Ali2
, Shireen R. Mohammed2 and Maher K. Ali2
Department of General Science, Collage of Basic Education, University of Duhok, Duhok, Iraq.
Department of Chemistry, Faculty of Science, University of Zakho, Zakho, Iraq.
Corresponding Author E-mail: layla.othman@uod.ac
DOI : http://dx.doi.org/10.13005/ojc/390214
Article Received on : 19 Dec 2022
Article Accepted on :
Article Published : 22 Mar 2023
Reviewed by: Prof. G. Rajitha
Second Review by: Dr. Manish tyagi
Final Approval by: Dr. Tanay Pramanik
The current assignment depicts the structure of recent quinoline derivatives. This method begins with the structure of imine derivatives through the condensation reaction of ethyl 2-aminobenzoate with various substituted aliphatic aldehydes and ketones in the existence of sodium hydroxide as a catalyst. While the second step includes the intra-cyclization process of the imine compounds in presence of a base like tertiary butoxide that resolute installs the hydroxyl- group on the bicyclic skeleton and aromatic amines. The molecular docking program Flare V4.0 was applied to investigate the biological activities of divers produced compounds against E.coli bacteria. Spectral data support the compounds of each the recent outputs acquired during this assignment.
KEYWORDS:Aliphatic Aldehyde and Ketone; Cyclization processes; Heterocyclic Compounds; Imine synthesis; Molecular Docking; Quinoline
Download this article as:| Copy the following to cite this article: Othman L. A, Mohammed S. R, Ali M. K. A Flexible Route to Synthesis and Molecular Docking of Some New Quinoline Derivatives through Imine and Cyclization Processes. Orient J Chem 2023;39(2). |
| Copy the following to cite this URL: Othman L. A, Mohammed S. R, Ali M. K. A Flexible Route to Synthesis and Molecular Docking of Some New Quinoline Derivatives through Imine and Cyclization Processes. Orient J Chem 2023;39(2). Available from: https://bit.ly/3JXgNkp |
Introduction
The heterocyclic structures include pronounced significance in pharmaceutical chemistry such as quinoline compounds that are established in more than two hundred surely existing alkaloids1-6. A wide diversity of quinoline showed a great range of biological activities such as antibiotic 7, antihypertensive 8, anti-HIV 9, tyro kinase PDGF-RTK inhibition10, Anticancer11, Antimalarial12, Anti-inflammatory13, antipsychotic14, anti-tuberculosis15, antifungal16, and antibacterial17. Traditional paths to quinolines core include Doebnere Miller and Friedlander18, Skraup19, and Combes synthesis strategies20,21. One of the simplest methods to get extremely functionalized quinoline derivatives in organic synthesis hold to be the Friedländer protocol, which prepared quinoline via mixing of o-amino benzaldehyde with acetaldehyde using sodium hydroxide as a catalyst. Accordingly, due to these considering full attention to the compositions and the biological activity of quinoline core, our present plan is the expansion of our attempts through styling, synthesis, and spotting of novel quinoline framework which forward via the condensation reaction of ethyl 2-aminobenzoate substrate with different aliphatic aldehydes and ketones, followed by the intra-cyclization process under basic conditions (Scheme 1).
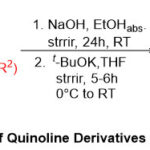 |
Scheme 1: Synthetic route of Quinoline Derivatives from Ethyl 2-Aminobenzoate. |
Result and Discussion
Steady quinoline products were achieved through two reaction steps 22. The reaction (Scheme 2) forwarded via imine construction by the condensation of ethyl 2-aminobenzoate 1 with various substituted of aliphatic aldehydes and ketones (2-9) in absolute ethanol, using NaOH as a catalyst with stirring for 24 hours at room temperature. In accordance with an effective purification step during twice recrystallization in absolute ethanol, pure expected products (10-17) were obtained with yields (65-75%) (Table 1). The FT-IR spectra showed a strong band for C=N imine group at νmax (1651-1373) cm-1. Respectively to the aromatic structure23, the 1HNMR spectrum of the products (10-17) has appeared new imine peaks at δ (8.05-7.82) ppm and extra proton peaks in the aliphatic region at δ (4.38-0.99) ppm. The occurrence of new C=N signals at δ (169.25-163.57) ppm, additional carbon peaks at δ (153.40-112.53) ppm for the aromatic and hetero aromatic regain, and (64.92-15.10) ppm of the aliphatic region of the 13CNMR spectra, confirming the structures of the expected products (10-17) 24.
 |
Scheme 2: Synthesis of Imines from Ethyl 2-aminobenzoate. |
Furthermore, the cyclization step of the imine products (10-17) (Scheme 3), forwarded with tBuOK in dry tetrahydrofuran. This procedure created the desired compounds (18-25) with yields (60-70%) (Table 2) after the purification method and using column chromatography . 1HNMR spectra of (18-25) appeared an additional number of proton peaks at δ (8.23-6.43) ppm of the aromatic and hetero aromatic regain, (11.20-10.00) ppm of OH, and (3.13-1.00) ppm of the alkyl group 25. The occurrence of new extra carbon peak at δ (176.28-105.00) ppm for the aromatic- and hetero-aromatic regain, and (35.30-14.99) ppm of the aliphatic regain in 13CNMR spectra confirming the structures of the desired products (18-25). In addition to, the molecular docking program was applied to the compounds shown in (Table 3) due to its high effectiveness.
 |
Scheme 3: Synthesis of Quinoline Derivatives from Imines. |
The proposed mechanism for the represented transformers of imines (10-17) to quinoline derivatives (18-25) is summarized in (Scheme 4). The first step involved proton removal from the imines (10-17) by tBuOK as a base at 0°C. Whilst at room temperature, the intermediates (10i-17i) undergo intra-cyclization (18i-25i), followed by aromatization (18-25) through the tautomerization step 26.
 |
Scheme 4: Mechanism of the Cyclization process of Imine Intermediates. |
 |
Table 1: Structures of Imine Products (10-17) from aliphatic aldehydes and Ketones (2-9). |
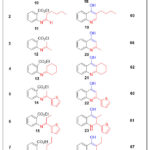 |
Table 2: Synthesis of Quinoline Derivatives (18-25) from Imines (10-17). |
 |
Table 3: Molecular Docking Result Data for Quinoline Derivatives |
 |
Figure 1: 2D and 3D Molecular Docking figure Contacting Compound 22 with Amino Acids of E.coli Bacteria’s RNA. Click here to View figure |
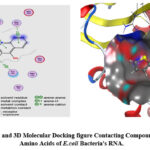 |
Figure 2: 2D and 3D Molecular Docking figure Contacting Compound 23 with Amino Acids of E.coli Bacteria’s RNA. |
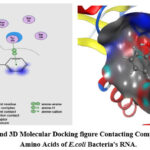 |
Figure 3: 2D and 3D Molecular Docking figure Contacting Compound 24 with Amino Acids of E.coli Bacteria’s RNA. |
 |
Figure 4: 2D and 3D Molecular Docking figure Contacting Compound 25 with Amino Acids of E.coli Bacteria’s RNA. |
Experimental Part
The melting points were registered via electro thermal apparatus and are not corrected. Each of reactions were achieved below the argon atmosphere with dry solvents below the dry conditions. 1HNMR and 13CNMR spectra were getting with DPX-300 Bruker, FT-NMR spectrometers at 500 and 125.8 respectively. On TLC 60 F254 Merck Kiese gel was placed on Mache Rey Nagel Aluminum foil. The detection was done using Ultraviolet light at 254-365 nanometers. The column chromatography was outright with Merck silica gel used for flash chromatography. The molecular docking program Flare V4.0 was applied to investigate the biological activities of divers produced compounds against E.coli bacteria.
General Protocol for preparation Imines (10-17) from Ethyl 2-aminobenzoate 1
To the solution of ethyl 2-aminobenzoate 1 (0.05 gm, 0.004 mmol, 1eq.) and aldehydes and ketones substituted (2-9) (0.004 mmol, 3 eq.) in ethanol absolute, a solution of NaOH (0.16 gm, 0.004 mmol, 1eq.) in ethanol absolute was added slowly with persistent stirring at 25°C for overnight. TLC (Pt/EtOAc 90:10) was used to observe the proceeding of the reaction. The crude product was obtained after evaporating the solvent below the vacuum followed by purification twice using ethanol absolute. On the basis of this process, the required products were received (10-17) 27 (Table 1).
Ethyl-2-(pentylideneamino) benzoate (10): (71%). (252-253)°C. FT-IR νmax (cm-1) = 3080, 2871, 2956, 1708, 1557, 1444. 1H NMR: δ(ppm) = 8.05 (t, 1H, CH=N), 7.94 (dd, 1H, CHaromatic), 7.48 (td, 1H, CHaromatic), 7.45 (dd, 1H, CHaromatic), 7.36 (td, 1H, CHaromatic), 4.31 (q, 2H, CH2), 2.40 (q, 2H, CH2), 1.58 (p, 2H, CH2), 1.41-1.36 (m, 5H, CH2), 0.99 (t, 3H, CH3). 13C NMR: δ(ppm) = 172.01 (C=O), 168.37 (C=N), 153.40 (Caromatic), 136.60 (Caromatic), 134.47 (Caromatic), 125.81 (Caromatic), 121.98 (Caromatic), 115.80 (Caromatic), 64.05 (C), 37.13 (C), 33.13 (C), 24.62 (C), 17.46 (C), 16.78 (C).
Ethyl-2-(hexylideneamino) benzoate (11): (70%). (122-123)°C. FT-IR νmax (cm-1) = 3050, 2859, 2955, 1708, 1595, 1453. 1H NMR: δ(ppm) = 7.82 (t, 1H, CH=N), 7.77 (dd, 1H, CHaromatic), 7.47 (td, 1H, CHaromatic), 7.39 (dd, 1H, CHaromatic), 7.34 (td, 1H, CHaromatic), 4.38 (q, 2H, CH2), 2.31 (q, 2H, CH2), 1.57-1.55 (m, 2H, CH2), 1.42-1.36 (m, 7H, CH2), 1.00 (t, 3H, CH3). 13C NMR: δ(ppm) = 171.97 (C=O), 168.33 (C=N), 153.35 (Caromatic), 136.56 (Caromatic), 134.42 (Caromatic), 125.77 (Caromatic), 121.94 (Caromatic), 115.75 (Caromatic), 64.00 (C), 36.69 (C), 33.73 (C), 29.87 (C), 25.65 (C), 17.41 (C), 16.73 (C).
Ethyl-2-(propan-2-ylideneamino) benzoate (12): (68%). (253-255) (Dec.)°C. FT-IR νmax(cm-1)= 3015, 3020, 2965, 1743, 1575, 1607, 1445, 1520. 1H NMR: δ (ppm) = 7.87 (dd, 1H,CHaromatic), 7.49-7.47 (m, 2H, CHaromatic), 7.37-7.35 (m, 1H, CHaromatic), 4.31 (q, 2H, CH2), 2.36 (s, 6H, CH3), 1.40 (t, 3H, CH3). 13C NMR: δ(ppm) = 174.42 (C=O), 169.25 (C=N), 152.28 (Caromatic), 137.61 (Caromatic), 135.01 (Caromatic), 125.23 (Caromatic), 121.90 (Caromatic), 114.83 (Caromatic), 64.92 (C), 31.00 (C), 27.28 (C), 18.33 (C).
Ethyl-2-(cyclohexylideneamino) benzoate (13): (70%). (275-276)°C. (cm-1)FT-IR νmax = 3010, 2888, 1746, 1578, 1448, 1519. 1H NMR: δ(ppm) = 7.96 (dd, 1H, CHaromatic), 7.5 (td, 1H, CHaromatic), 7.44 (dd, 1H, CHaromatic), 7.37 (td, 1H, CHaromatic), 4.33 (q, 2H, CH2), 2.55 (t, 4H, CH2), 1.71 (p, 4H, CH2), 1.46 (p, 2H, CH2), 1.38 (t, 3H, CH3). 13C NMR: δ(ppm) = 181.73 (C=O), 166.33 (C=N), 149.08 (Caromatic), 134.53 (Caromatic), 131.99 (Caromatic), 123.79 (Caromatic), 119.63 (Caromatic), 112.53 (Caromatic), 62.00 (C), 37.29 (2C), 33.52 (C), 27.00 (C), 25.75 (C), 15.41 (C).
Ethyl-2-((1-(thiophen-2-yl) ethylidene) amino) benzoate (14): (72%). (287-288) (Dec.)°C. FT-IR νmax (cm-1)= 3074, 2929, 2975, 1743, 1547, 1612, 1447. 1H NMR: δ(ppm) = 7.98 (dd, 1H, CHaromatic), 7.53-7.43 (m, 3H, CHaromatic), 7.38-7.35 (m, 2H, CHaromatic), 7.20 (t, 1H, CHar), 4.31 (q, 2H, CH2), 2.60 (s, 3H, CH3), 1.39 (t, 3H, CH3). 13C NMR: δ(ppm) = 167.33 (C=O), 163.57 (C=N), 147.65 (Caromatic), 146.61 (Caromatic), 135.64 (Caromatic), 133.00 (Caromatic), 131.77 (Caromatic), 129.13 (Caromatic), 126.30 (Caromatic), 125.40 (Caromatic), 120.33 (Caromatic), 114.29 (Caromatic), 63.00 (C), 27.06 (C), 16.41 (C).
Ethyl-2-((1-(thiophen-2-yl) propylidene) amino) benzoate (15): (75%). (298-299) (Dec.)°C. FT-IR νmax(cm-1)= 3015, 2970, 1725, 1575, 1651, 1485, 1522. 1H NMR: δ(ppm) = 7.98 (dd, 1H, CHaromatic), 7.52-7.49 (m, 3H, CHaromatic), 7.39-7.35 (m, 2H, CHaromatic), 7.19 (t, 1H, CHaromatic), 4.31 (q, 2H, CH2), 2.79 (q, 2H, CH2), 1.53 (t, 3H, CH3), 1.39 (t, 3H, CH3). 13C NMR: δ(ppm) = 169.15 (C=O), 165.69 (C=N), 149.60 (Caromatic), 148.39 (Caromatic), 137.36 (Caromatic), 134.81 (Caromatic), 133.59 (Caromatic), 132.18 (Caromatic), 130.95 (Caromatic), 127.92 (Caromatic), 122.70 (Caromatic), 116.38 (Caromatic), 64.82 (C), 34.86 (C), 18.24 (C), 15.10 (C).
Ethyl-2-((1-phenylbutylidene) amino) benzoate (16): (70%). (265-266) (Dec.)°C. FT-IR νmax(cm-1) = 3029, 2980, 1742, 1578, 1608, 1427, 1520. 1H NMR: δ(ppm) = 7.87 (dd, 1H, CHaromatic), 7.62 (dd, 2H, CHaromatic), 7.50-7.47 (m, 5H, CHaromatic), 7.37 (td, 1H, CHaromatic), 4.34 (q, 2H, CH2), 2.35 (t, 2H, CH2), 1.68-1.64 (m, 2H, CH2), 1.40 (t, 3H, CH3), 1.03 (t, 3H, CH3). 13C NMR: δ(ppm) = 168.37 (C=O), 167.35 (C=N), 148.87 (Caromatic), 137.03 (Caromatic), 135.57 (2Caromatic), 133.20 (Caromatic), 133.01 (Caromatic), 130.46 (2Caromatic), 129.45 (Caromatic), 126.12 (Caromatic), 120.90 (Caromatic), 114.58 (Caromatic), 63.03 (C), 39.53 (C), 22.10 (C), 16.44 (C), 15.31 (C).
Ethyl-2-((3-phenylbutylidene) amino) benzoate (17): (70%). (99-101)°C. FT-IR νmax (cm-1) = 3059, 3025, 2963, 2828, 1745, 1601, 1560, 1451. 1H NMR: δ(ppm) = 7.92 (t, 1H, CH=N), 7.65 (dd, 1H, CHaromatic), 7.48 (td, 1H, CHaromatic), 7.42 (dd, 1H, CHaromatic), 7.37 (td, 1H, CHaromatic), 7.18-7.14 (m, 5H, CHaromatic), 4.37 (q, 2H, CH2), 2.87-2.81 (m, 1H, CH2), 2.63-2.58 (m, 1H, CH), 2.43-2.38 (m, 1H, CH3), 1.39-1.36 (m, 6H, CH3). 13C NMR: δ(ppm) = 171.98 (Caromatic), 168.86 (Caromatic), 156.77 (Caromatic), 149.65 (Caromatic), 137.09 (Caromatic), 134.96 (2Caromatic), 131.52 (2Caromatic), 130.62 (Caromatic), 129.82 (Caromatic), 126.31 (Caromatic), 122.48 (Caromatic), 116.29 (Caromatic), 64.54 (C), 44.29 (C), 43.50 (C), 23.28 (C), 17.95 (C).
General Protocol for Preparation of Quinoline Derivatives (18-25) from Imines (10-17):
To a mixture of imines (10-17) (0.173 mmol, 1 eq.) in dry tetra hydro furan (THF) (10 ml), tBuOK (tertiary butoxide) (0.05 gm, 0.458 mmole, 2.65 eq.) as added in one portion at 0°C with stirring (5-6 h) at 25°C. The reaction mixture was subsides with a few drops of an NH4Cl solution. The dissolvent was evaporated below the vacuum. The remains were extracted with DCM, followed by washing with salt solution and drying through magnesium sulfate. The solvent was concentrated with lower pressure succeed by purification through the flash chromatography to afford the desired products (18-25) 28,29(Table 2).
3-propylquinolin-4-ol (18): (60%). (86-88)°C. FT-IR νmax(cm-1) = 3428, 3400, 3030, 2956, 1590, 1457. 1H NMR: δ(ppm) = 11.04 (s, 1H, OH), 8.17 (s, 1H, CH=N), 7.90 (dd, 1H, CHaromatic), 7.80 (dd, 1H, CHaromatic), 7.50 (td, 1H, CHaromatic), 7.32 (td, 1H, CHaromatic), 2.46 (t, 2H, CH2), 1.80-1.73 (m, 2H, CH2), 1.05 (t, 3H, CH3). 13C NMR: δ (ppm) = 167.28 (Caromatic), 152.79 (Caromatic), 144.53 (Caromatic), 132.15 (Caromatic), 131.25 (Caromatic), 128.90 (Caromatic), 128.30 (Caromatic), 127.27 (Caromatic), 127.13 (Caromatic), 33.14 (C), 26.81 (C), 15.57 (C).
3-butylquinolin-4-ol (19): (65%). (97-99)°C. FT-IR νmax (cm-1) = 3365, 3295, 3035, 2928, 2955, 1607, 1458. 1H NMR: δ(ppm) = 11.21 (s, 1H, OH), 8.23 (s, 1H, CH=N), 7.88 (dd, 1H, CHaromatic), 7.85 (dd, 1H, CHaromatic), 7.50 (td, 1H, CHaromatic), 7.34 (td, 1H, CHaromatic), 2.61 (t, 2H, CH2), 1.72 (p, 2H, CH2), 1.44-1.40 (m, 2H, CH2), 1.00 (t, 3H, CH3). 13C NMR: δ(ppm) = 166.06 (Caromatic), 151.04 (Caromatic), 142.54 (Caromatic), 130.53 (Caromatic), 129.62 (Caromatic), 126.76 (Caromatic), 126.68 (Caromatic), 125.65 (Caromatic), 124.74 (Caromatic), 32.57 (C), 32.03 (C), 22.45 (C), 14.99 (C).
2-methylquinolin-4-ol (20): (62%). (234-235) (Dec.)°C. IR νmax (cm-1) = 3388, 3300, 3040, 2960, 1601, 1485. 1H NMR: δ(ppm) = 10.57 (s, 1H, OH), 7.95 (dd, 1H, CHaromatic), 7.83 (dd, 1H, CHaromatic), 7.50 (td, 1H, CHaromatic), 7.30 (td, 1H, CHaromatic), 6.43 (s, 1H, CHaromatic), 2.65 (s, 3H, CH3). 13C NMR: δ(ppm) = 168.61 (Caromatic), 160.21 (Caromatic), 143.77 (Caromatic), 130.16 (Caromatic), 127.04 (Caromatic), 124.19 (Caromatic), 122.94 (Caromatic), 116.44 (Caromatic), 110.67 (Caromatic), 23.40 (C).
1, 2, 3, 4-tetrahydroacridin-9-ol (21): (60%). (133-135)°C. FT-IR νmax (cm-1) = 3377, 3365, 3119, 2988, 1625, 1399. 1H NMR: δ(ppm) = 11.03 (s, 1H, OH), 7.88 (dd, 1H, CHaromatic), 7.78 (dd, 1H, CHaromatic), 7.45 (td, 1H, CHaromatic), 7.28 (td, 1H, CHaromatic), 3.13 (t, 2H, CH2), 2.81 (t, 2H, CH2), 1.79-1.72 (m, 4H, CH2). 13C NMR: δ(ppm) = 167.90 (Caromatic), 165.03 (Caromatic), 142.03 (Caromatic), 134.05 (Caromatic), 131.85 (Caromatic), 128.20 (Caromatic), 127.40 (Caromatic), 124.76 (Caromatic), 120.38 (Caromatic), 35.30 (C), 30.39 (C), 27.65 (C), 26.67 (C).
2-(thiophen-2-yl) quinolin-4-ol (22): (61%). (115-117)°C. FT-IR νmax(cm-1) = 3360, 3325, 3100, 2922, 1610, 1454. 1H NMR: δ(ppm) = 10.00 (s, 1H, OH), 8.12 (dd, 1H, CHaromatic), 7.95 (dd, 1H, CHaromatic), 7.81 (dd, 1H, CHaromatic), 7.58 (td, 1H, CHaromatic), 7.46 (dd, 1H, CHaromatic), 7.40 (td, 1H, CHaromatic), 7.32 (t, 1H, CHaromatic), 6.96 (s, 1H, CHaromatic). 13C NMR: δ(ppm) = 176.28 (Caromatic), 161.54 (Caromatic), 151.11 (Caromatic), 141.21 (Caromatic), 136.66 (Caromatic), 136.13 (Caromatic), 134.40 (Caromatic), 133.44 (Caromatic), 131.80 (Caromatic), 129.27 (Caromatic), 126.28 (Caromatic), 122.23 (Caromatic), 105.00 (Caromatic).
3-methyl-2-(thiophen-2-yl)-1,2-dihydroquinolin-4-ol (23): (67%). (92-93)°C. FT-IR νmax(cm-1) = 3355, 3325, 3090, 2922, 1606, 1455. 1H NMR: δ(ppm) = 10.60 (s, 1H, OH), 7.88 (dd, 1H, CHaromatic), 7.81 (dd, 1H, CHaromatic), 7.60 (dd, 1H, CHaromatic), 7.48 (td, 1H, CHaromatic), 7.29-7.23 (m, 3H, CHaromatic), 2.03 (s, 3H, CH3). 13C NMR: δ(ppm) = 174.14 (Caromatic), 159.49 (Caromatic), 147.39 (Caromatic), 145.57 (Caromatic), 137.08 (Caromatic), 136.01 (Caromatic), 134.87 (Caromatic), 133.85 (Caromatic), 132.11 (Caromatic), 131.87 (Caromatic), 130.09 (Caromatic), 121.54 (Caromatic), 119.32 (Caromatic), 15.27 (C).
3-ethyl-2-phenylquinolin-4-ol (24): (65%). (120-121)°C. FT-IR νmax(cm-1) = 3426, 3420, 3040, 2850, 1633, 1430. 1H NMR: δ(ppm) = 11.05 (s, 1H, OH), 7.95 (td, 2H, CHaromatic), 7.89 (dd, 2H, CHaromatic), 7.53 (td, 1H, CHaromatic), 7.44 (t, 2H, CHaromatic), 7.38-7.35 (m, 2H, CHaromatic), 2.82 (q, 2H, CH2), 1.31 (t, 3H, CH3). 13C NMR: δ(ppm) = 170.64 (Caromatic), 160.06 (Caromatic), 142.12 (Caromatic), 136.54 (Caromatic), 132.02 (Caromatic), 131.98 (Caromatic), 130.95 (Caromatic), 129.96 (Caromatic), 129.70 (2Caromatic), 127.86 (Caromatic), 126.11 (Caromatic), 123.35 (2Caromatic), 120.04 (Caromatic), 24.19 (C), 15.00 (C).
3-(1-phenylethyl) quinolin-4-ol (25): (60%). (78-80)°C. FT-IR νmax (cm-1) = 3330, 3299, 3028, 2960, 1601, 1485. 1H NMR: δ (ppm) = 8.29 (s, 1H, OH), 7.88 (dd, 1H, CHaromatic), 7.86 (dd, 1H, CHaromatic), 7.51 (td, 1H, CHaromatic), 7.35 (td, 1H, CHaromatic), 7.29-7.23 (m, 5H, CHaromatic), 7.21-7.18 (m, 1H, CHaromatic), 4.16 (q, 1H, CH), 1.63 (d, 3H, CH3). 13C NMR: δ(ppm) = 171.49 (Caromatic), 151.64 (Caromatic), 150.72 (Caromatic), 146.44 (Caromatic), 133.82 (2Caromatic), 132.52 (Caromatic), 132.42 (Caromatic), 132.26 (Caromatic), 130.06 (2Caromatic), 130.02 (Caromatic), 128.28 (Caromatic), 128.18 (Caromatic), 127.48 (Caromatic), 42.60 (C), 27.84 (C).
Molecular Docking Study
The molecular docking program Flare V 4.0 30,31 was performed for all recently synthesized quinoline derivatives 22-25 with the crystallographic structure of Escherichia coli bacteria (E.coli) ID: 1NYM as a target molecule. The aim of molecular docking is to give a prediction of the ligand-receptor complex structure using computation methods. The docking process involves two basic steps: prediction of the ligand conformation as well as its position and orientation within these sites (usually referred to as pose) and assessment of the binding affinity. Docking is generally used to determine the best binding direction of molecules linked to a protein molecule in order to predict binding energy and biological activity. The active point of the receptor (E.coli) contains the amino acid residues ASN 132, ASN 170, GLU 104, GLU 240, SER 70, SER 130, TYR 105, ALA 237, GLY 238, GLY 236, ARG 244, VAL 216, SER 235 32. It was discovered that all of the compounds share interactions with the amino acids GLU 240, SER 130, ALA 237, GLY 238, VAL 216, and SER 235 generally. The ligand interaction graphs of the synthesized compounds were shown in figures 1, 2, 3, and 4 to clarify the potential for inhibitory activity. Table 3 shows the docking scores of the investigated compounds. The docking program reveals that all compounds 22-25 bind with the active center of amino acid of Escherichia coli bacteria by several types of bonds, such as: (Vander Waal force, carbon-hydrogen bond, π-Anion, π-Alkyl, and the most important one, hydrogen bond). The most critical interactions between ligands and the active point of bacteria’s RNA amino acids are hydrogen bond interactions. The docking scores for all compounds were good, ranging from -10.18 kilojoules per pole to -11.02 kilojoules per mole. The molecular docking revealed that the best direction of compounds in the binding pocket was created by hydrogen bonds formed by the OH groups of the ligand with amino acid of ASN 132, SER 235, VAL 216, TYR 105 and ALA 237, figure 4. Resulting in an increase in the negativity of ΔG values, as shown in Table3. The increase of ΔG values negativity means that the compound has high biological activity due to its strong connection to the target bacteria’s RNA amino acids. While compound 24, figure 3. Connects only by the hydrogen bonds with the ARG 244, and VAL 216 amino acids of the RNA of the target bacteria this leads to having the lowest biological activity among all products due to having the lowest ΔG negativity values Table 3.
Conclusion
Briefly, suitable and useful formation of quinoline derivatives is sketched. The procedure was forwarded firstly with imine synthesis through the condensation reaction between aliphatic aldehydes and ketones with ethyl 2-aminobenzoate substrate under basic and soft conditions, followed by the functional intramolecular cyclization process to furnish the desired quinoline derivatives products in moderate yields. Ultimately, straightforwardly obtained products, flexible reaction conditions, and stubby reaction times are the fundamental utility of these processes. Furthermore, using the molecular docking program, all of these compounds showed good antibacterial action against E.coli bacteria.
Acknowledgement
Authors are thankful to the Chemistry Department, Faculty of Science, Zakho University for provide all facilities.
References
- Mhaske, S.B., Argade, N.P. The chemistry of recently isolated naturally occurring quinazolinone alkaloids, Tetrahedron. 2006, 62, 9787-9826.
CrossRef - Mohammed, S., Khalid, M. Aflexible synthesis of naphthyridine derivatives through diazotization, triflation, and Suzuki reaction, Indian J. Heterocycl. Chem. 2019, 29, 21-25.
CrossRef - Mohammed, S., Maher, K. A facile entry to fused dipyrimidine: Preparation of imidazo [1, 2-a: 3, 4-a’] dipyrimidine-4, 9 (3H)-dione and pyrimido [1’, 2’:4, 5] pyrazino [1, 2-a] pyrimidine-4, 10 (3H, 6H)-dione derivatives, Indian J. Heterocycl. Chem. 2017, 27, 289-294.
CrossRef - Mohammed, S., Maher, K. Synthesis and spectral characterization of 1, 5-naphthyridine derivatives through cross-coupling Suzuki reaction, Indian J. Heterocycl. Chem. 2019, 29, 199-203.
CrossRef - Dawood, A.A., Mohammed, S.R., Mahmoud, M. Synthesis, identification and biological activity of new heterocyclic compounds from reaction of new schiff-bases with phathalic anhydride, Sci. J. Univ. Zakho. 2020, 8, 12-18.
CrossRef - Mohammed, S., Maher, K. AFacile Synthesis of Quinazolinone derivatives through vilismeierintermediate. Indian J. Heterocycl. Chem. 2017, 27, 83-87.
- Bénard, C., Zouhiri, F., Normand-Bayle, M., Danet, M., Desmaële, D., Leh, H., & d’Angelo, J. Linker-modified quinoline derivatives targeting HIV-1 integrase: synthesis and biological activity. Bioorganic & medicinal chemistry letters. 2004, 14(10), 2473-2476.
CrossRef - Maguire, M. P., Sheets, K. R., McVety, K., Spada, A. P., & Zilberstein, A. A new series of PDGF receptor tyrosine kinase inhibitors: 3-substituted quinoline derivatives. Journal of medicinal chemistry. 1994, 37(14), 2129-2137.
CrossRef - Solomon,V.R.,Lee, H., Quinoline as a privileged scaffold in cancer drug discovery, Curr.Med. Chem. 2011, 18, 1488-1508, doi: 10.2174/092986711795328382.
CrossRef - Kumar, A., Paliwal, D., Saini, D., Thakur, A., Aggarwal, S., & Kaushik, D. A comprehensive review on synthetic approach for antimalarial agents. European Journal of Medicinal Chemistry. 2014, 85, 147-178.
CrossRef - El-Feky, S. A., Abd El-Samii, Z. K., Osman, N. A., Lashine, J., Kamel, M. A., & Thabet, H. K. Synthesis, molecular docking and anti-inflammatory screening of novel quinoline incorporated pyrazole derivatives using the Pfitzinger reaction II. Bioorganic chemistry. 2015, 58, 104-116.
CrossRef - Zajdel, P., Partyka, A., Marciniec, K., Bojarski, A. J., Pawlowski, M., & Wesolowska, A. Quinoline-and isoquinoline-sulfonamide analogs of aripiprazole: novel antipsychotic agents? Future medicinal chemistry. 2014, 6(1), 57-75.
CrossRef - Lin, Y. M., Zhou, Y., Flavin, M. T., Zhou, L. M., Nie, W., & Chen, F. C. Chalcones and flavonoids as anti-tuberculosis agents. Bioorganic & medicinal chemistry. 2002, 10(8), 2795-2802.
CrossRef - Lahtchev, K. L., Batovska, D. I., Parushev, S. P., Ubiyvovk, V. M., & Sibirny, A. A. In vitro antifungal evaluation and structure-activity relationships of a new series of chalcone derivatives and synthetic analogues, with inhibitory properties against polymers of the fungal cell wall. Eur J Med Chem. 2008, 43(10), 2220-2228.
CrossRef - Sivakumar, P. M., Ganesan, S., Veluchamy, P., & Doble, M. Novel chalcones and 1, 3, 5‐triphenyl‐2‐pyrazoline derivatives as antibacterial agents. Chemical biology & drug design. 2010, 76(5), 407-411.
CrossRef - Yakes, F. M., Chen, J., Tan, J., Yamaguchi, K., Shi, Y., Yu, P., & Joly, A. H. Cabozantinib (XL184), a novel MET and VEGFR2 inhibitor, simultaneously suppresses metastasis, angiogenesis, and tumor growth. Molecular cancer therapeutics. 2011,10(12), 2298-2308.
CrossRef - Porter, J.Small molecule c-Met kinase inhibitors: a review of recent patents. Expert opinion on therapeutic patents. 2010,20(2), 159-177.
CrossRef - Friedländer, P., & Weinberg, A. Ueber einige im Pyridinkern substituirte Chinolinderivate. Berichte der deutschen chemischen Gesellschaft. 1882, 15(2), 2679-2685.
CrossRef - Skraup, Z. H. Eine Synthesis des Chinolins Ber. 1880, 13, 2086.
CrossRef - Combes, A. Quinoline Synthesis. Bull. Soc. Chim, Fr.1888, 49, 89.
- Joule, J. A., Mills, K., & Smith, G. F. Heterocyclic Chemistry. CRC Press, 2020.
CrossRef - Othman, L. A., Mohammed, S. R., & Khalid, M. Synthesis and Characterization of Some New Quinoline Derivatives Derived from 2-Amino Benzonitrile. Indian Journal of Heterocyclic Chemistry. 2022, 32(04), 487-492.
- Al-Hasani RAM. Preparation, structural and antimicrobial studies of a new bimetallic complexes involving a new schiff and mannich bases. Al-Nahrain J Sci. 2007,10(2):39-49.
CrossRef - Bhat MA, Al-Omar MA. Synthesis, characterization and in vivo anticonvulsant and neurotoxicity screening of Schiff bases of phthalimide. Acta Pol Pharm. 2011, 68(3):375-380.
- Bakkar, M., Monshi, M., Warad, I., Siddiqui, M., & Bahajaj, A. 1H 13C NMR investigation of E/Z-isomerization around CN bond in the trans-alkene-Pt (II) imine complexes of some ketimines and aldimines. Journal of Saudi Chemical Society. 2010, 14(2), 165-174.
CrossRef - Mohammed, S., & Khalid, M. A. H. E. R. A Facile Protocol for the Construction of Tricyclic Framework Tetrahydrobenzo-4-nitrobenzenesulfonate, 4-methylbenzenesulfonate and [1, 8] naphthyridine Substituents from Methyl δ-Lactam. Orient. J. Chem. 2015, 31(4), 2137-2146.
CrossRef - Azzouz, A. S. P., & Ali, R. T. Synthesis of Schiff bases derived from benzaldehyde and salicylaldehyde with some amino acids by a new develop method. National Journal of Chemistry. 2010, 37, 158-168.
CrossRef - Mohammed, S.R. Development of New Radical Processes: Approaches toward the Synthesis of Eucophylline, Institut des Sciences Moléculaires, France. 2014.
- Hassan, H., Mohammed, S., Robert, F., Landais, Y. Total synthesis of (±)-eucophylline. A free-radical approach to the synthesis of the azabicyclo [3.3.1] nonane skeleton, Org. Lett. 2015, 17, 4518-4521.
CrossRef - Milo, S., Heylen, R. A., Glancy, J., Williams, G. T., Patenall, B. L., Hathaway, H. J. & Jenkins, A. T. A. A small-molecular inhibitor against Proteus mirabilis urease to treat catheter-associated urinary tract infections. Scientific reports, 2021. 11(1), 1-15.
CrossRef - Yoshino, M., & Murakami, K. A graphical method for determining inhibition constants. Journal of Enzyme Inhibition and Medicinal Chemistry. 2009, 24(6), 1288-1290.
CrossRef - Mesibov, R., & Adler, J. Chemotaxis toward amino acids in Escherichia coli. Journal of Bacteriology.1972, 112(1), 315-326.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.









