Method Development and Validation for Estimation of Cefadroxil in Different Marketed Tablets by UV Spectrophotometry Method and Anti-Inflammatory Studies Using In-Silico Approaches
Department of Pharmaceutics, Sanskar College of Pharmacy and Research (SCPR), Sanskar Educational Group, Ghaziabad, Uttar Pradesh, India.
Corresponding Author E-mail: shabnam.ain@sanskar.org
DOI : http://dx.doi.org/10.13005/ojc/380409
Article Received on : 14 Mar 2022
Article Accepted on :
Article Published : 06 Jul 2022
Reviewed by: Dr. Nitin Rawat
Second Review by: Dr. Atul Goswami
Final Approval by: Dr. Amira Cipurkovic
Quality-based assessment of pharmaceuticals obviates the uncertainties concerning their quality, safety and efficacy for their regulatory purpose. A method was developed and validated for quality control assessment of cefadroxil for the pharmaceuticals or row material analysis. In-silico analysis wasperformedto evaluate the bioavailability, toxicity as well asanti-inflammatory potential of cefadroxil. The results showed that the developed method was found linear, accurate, precise and robust while the dissolution rate of each tablet was found comparable. In-silico docking analysis and network pharmacology analysis showed low bioavailability and toxicity as well as a significant anti-inflammatory potential of cefadroxil via regulation of genes such as TNF-α, IL-6, SLC15A1 and SLC15A2. However, due to its bioavailability barriers, further experimental strategies are necessary to re-purpose the therapeutic application of cefadroxil as a potent anti-inflammatory agent.
KEYWORDS:Anti-Inflammatory Activity; Bioavailability; Cefadroxil; Method validation
Download this article as:| Copy the following to cite this article: Ali Z, Ain S, Kumar B, Ain Q. Method Development and Validation for Estimation of Cefadroxil in Different Marketed Tablets by UV Spectrophotometry Method and Anti-Inflammatory Studies Using In-Silico Approaches. Orient J Chem 2022;38(4). |
| Copy the following to cite this URL: Ali Z, Ain S, Kumar B, Ain Q. Method Development and Validation for Estimation of Cefadroxil in Different Marketed Tablets by UV Spectrophotometry Method and Anti-Inflammatory Studies Using In-Silico Approaches. Orient J Chem 2022;38(4). Available from: https://bit.ly/3yLQb04 |
Introduction
Cephalosporin antibiotics have been associated with a tremendous impact on the treatment of infectious diseases, clinically. Cephalosporins and the semisynthetic penicillins are closely related in the structure; all contain a β-lactam ring and a dihydrothiazide ring which includes sulphur. The isolation of the active component of cephalosporin, C-7-aminocephalosporanic acid, made possible the synthesis of different compounds whose varying side chains impart antibacterial activity and pharmacokinetic properties. Since, the history of oral antimicrobial drugs, the cephalosporins are one of the categorized anti-bacterial and anti-inflammatory drugs which produced a large group of highly effective bactericidal compounds that are widely applicable in the treatment of bacterial infections, especially in pediatric patients. These drugs hyphenated many of the necessary characteristics of oral antimicrobials 1.
Cefadroxil is one of the famous antibiotics used to treat infection caused by gram-positive and gram-negative microorganisms namely,Enterococcus faecalis, Escherichia coli, Bacteroides fragilis, Pseudomonas aeruginosa, Staphylococcus pseudintermedius, Staphylococcus aureus, Streptococcus pneumoniaetc2.Cefadroxil falls under first-generation antibiotics of the cephalosporin class that inhibits the microbial cell wall synthesis and that is why these are acknowledged asa bactericidal antibiotic. The United States Food and Drug Administration characterized cefadroxilas especially to treat the cutaneous, urinary and respiratory tract infection and other systemic bacterial infections caused by staphylococci 3. The cefadroxil ought to be warily practiced to the patients associated with extreme hypersensitivity topenicillin because of its structural similarity. Moreover, physicochemical characteristics of cefadroxil incorporate, whitish-yellow translucent powder, the solvent in water and stability in acidic conditions. The cefadroxil was reported to exhibit a sophisticated half-life along with sustained efficacy than other drug molecules of the same class45.
Regulatory-based assessment of pharmaceuticals is critically needed to validate them based on their quality, safety, and efficacy purpose. Moreover, various analytical techniques are highly associated with the analysis of drugs, qualitatively and quantitatively. The most used techniques for the analysis of drugs are namely UV-Visible spectroscopy, chemiluminescence, near-infrared spectroscopy, potentiometry, polarography, High-Performance Liquid Chromatography (HPLC), gel filtration chromatography, High-Performance Thin Layer Chromatography (HPTLC) capillary zone electrophoresis, Liquid Chromatography and Mass spectroscopy (LC-MS)and Nuclear Magnetic Resonance Spectroscopy (NMR) methods 6–8. UV visible spectroscopy is one of the highly abundant, most used, and economic analytical techniques for qualitative and quantitative standardization of drugs or pharmaceuticals. Several biological approaches are used to evaluate the therapeutic and associated negative effects of a single drug or newly developed pharmaceuticals 9. Furthermore, in-silico approaches put an important contribution to evaluating the therapeutic potential of drug molecules based on ligation strength with the specific targeted gene. Besides, network pharmacology analysis uncovers the multi-mechanistic and therapeutic approach of different tested drugs or active pharmaceutical ingredients 10,11.
Considering the
above facts, the study is aimed to develop and validate a method for the
estimation of cefadroxil and to explore the multi-mechanistic role of
cefadroxil in inflammation using computational approaches.
Material and Methods
Chemicals, Reagents and Softwares
Different brands of cefadroxil (500 mg) tablets such as Odoxil (T1), Bicef (T2), Cefadrox (T3), and Droxyl (T4) were purchased from the local retail pharmacy, sucrose, glycerine and sorbitol (SRL chemicals Pvt. Ltd. India), double distilled water, SwissADME tool, Cytoscape (Version 3.8.2), Autodock Vina (Version 1.5.7).
Collection of cefadroxil tablets
The tablets of cefadroxil of different brands were procured from the local retail pharmacy store and kept for further analysis. The production company name, marketed company name, batch number, date of manufacturing and date of expiry was recorded for record purpose.
Determination of absorption maxima and method validation for quantitative analysis of cefadroxil in different tablets
One milligram (mg) of cefadroxil was dissolved in the double-distilled water with the help of a vortex for 10 min so that the content of the drug could be dissolved properly. After vortex, the mixture was centrifuged for 5 minutes (min) at 10000 rpm and the obtained supernatant was separated and analysed spectrophotometrically to determine the absorption maxima of cefadroxil. The measurements were recorded in triplicate.In quantitative analysis of cefadroxil, validation analysis was performed against the different concentrationsof cefadroxil ranging from 05-30 µg/ml were prepared in water. A calibration plot was generated to predict the linearity range of the developed method using UV spectrophotometer. The analysis was performed as per International Council on Harmonisation (ICH) guideline. The analysis was constrained to the linearity, the limit of detection (LOD), the limit of quantification (LOQ), precision, accuracy and robustness of the developed method.
Preparation of sample solution for quantitative analysis of cefadroxil in different tablets
Briefly, one tablet from each brand was crushed and dissolved in the water. The vortex method was used for the proper dissolution of the tablet drug content. The obtained mixtures were centrifuged at 10000 rpm for 10 min. The supernatant was separated from the vial and proceeded for the quantitative analysis spectrophotometrically. Each measurement was carried out in triplicate and the average reading or mean value was recorded for final statistical analysis.
Determination of dissolution rate of different tablets
Dissolution analysiswas carried out for different tablets under the defined condition as per protocol. Media assortmentwas based on the dissolution test purpose via considering the solubility of the drug.The dissolution test for different cefadroxil tablets was determined as per standard protocol as described in reported literature or United States Pharmacopeia. Briefly, a tablet of the different brands was placed in the basket/apparatus containing dissolution media with the constant rotation per minute (rpm) which is 50 rpm. The dissolution efficiency was carried out on each fraction/test sample withdrawn from the basket on 0, 2, 5, 8, 10, 15, 20, 30, and 45 minutes. The dissolution rate of cefadroxil was determined using UV spectrometry methods at 232 nm. Each measurement was carried out in triplicate12.
A standard plot of cefadroxil was plotted in the concentration ranging from 05 to 30 μg/ml. the volume of each concentration was made up ofthe tested dissolution medium. The studywas performed as per the International Conference of Harmonization (ICH) and the United States Pharmacopoeia, USP guideline. A blank formulation was also comprised by concocting a solution made up ofglycerine (1%),sucrose (1%), and sorbitol (1%) in water to evaluate the significance level via interferences analysis.
ADME/TOX analysis
ADME (absorption, distribution, metabolism and excretion) and toxicological examination was determined for cefadroxil through “SwissADME (http://www.swissadme.ch/index.php)” and ProTox-II- chemicals toxicityPrediction tool(https://tox-new.charite.de/protox_II/index.php?site=home). TPSA (Topological Polar Surface Area (TPSA) for drug property, lipophilicity as for drug Consensus Log Po/w, skin permeation as Log Kp, and drug-likeness was predicted in ADME response of cefadroxil 13.
In-silico analysis for determination of anti-inflammatory activity of cefadroxil
Autodock analysis for the anti-inflammatory activity of cefadroxil
Accession of the target protein
The three-dimensional structure of the targeted protein TNF-α and IL-6 was found in RCSB Protein Data Bank (http://www.rcsb.org/pdb; ID-6rmj and 1n26, respectively).
Ligand preparation
The ligands in SDF format weretransferred from the database of PubChem (https://pubchem.ncbi.nlm.nih.gov/compound/47965) and formatted in PDB and PDBQT format using software (BIOVIA Discovery Studio Visualizer 2021) and further processed for Autodock for molecular docking by adjusting torsion, ionization, degree of freedom and stereo-chemical variation 14.
Protein structure for docking
The Auto dock was used to prepare and refine the selected protein structure (TNF-α and IL-6)15. The protein structure in PDB format was downloaded with 1.7 A0 resolution and 0.222 and 0.171R-value free and R-value work, respectively. Molecular docking was performed by Auto dock Vian and BIOVIA Discovery Studio Visualizer software. Further, the processing of the docking analysis was performed using command Prompt and prerequisite before the docking analysis16.
Table 1: In-silico docking analysis of cefadroxil against TNF-α and IL-6.
|
Proteins name |
Binding Site |
Center Grid box |
Binding energy (kcal/mol) |
Interaction (conventional hydrogen bond; Donor and Acceptor) |
|
TNF-α |
Site 1 |
X-15.213, Y-41.124, Z-102.428 |
-6.8 |
ALA B:156, PRO B:12, GLU B:53, ASP B:10, LEU B:55, SER C: 9 |
|
Site 2 |
X-(-1.165), Y-67.434, Z-131.209 |
-6.7 |
ASP A:140, GLY A:24, LEU A:142 |
|
|
Site 3 |
X-12.131, Y-48.237, Z-91.177 |
-6.7 |
ASP A:140, GLY A:24, LEU A:142 |
|
|
IL-6 |
Site 1 |
X-16.234, Y-59.231, Z-75.632 |
-6.4 |
PRO A:289, ALA A:291, ILE A:203 |
|
Site 2 |
X-15.077, Y-32.959, Z-104.342 |
-6.8 |
ALA A:291, PRO A:289, PRO A:200, ILE A: 203, |
|
|
Site 3 |
X-43.264, Y-55.591, Z-44.16 |
-6.8 |
ARG A:82, GLY A:85, CYS A:77, THR A:86, TYR A:78, SER A:76, ARG A:44, |
Network pharmacology analysis for anti-inflammatory analysis of cefadroxil
Different targets or proteins or genomes were screened from Genecard (https://www.genecards.org/), Uni Port ID of each target was obtained Uni Port database (https://www.UniProt.org/uploadlists/) 11. The analysis was performed based on the gene-gene or gene-compound ligation efficacy. Compound-proteins interactions were generated using Cytoscape (version 3.8.2) software for interface evidence of each interacted gene with cefadroxil through integration analysis. The study enclosed all the genes which possessed nearly functional interactions with cefadroxil11,17.
Results and Discussion
Determination of absorption maxima and method validation for quantitative analysis of cefadroxil in different tablets
The absorption maxima was examined using the UV spectrophotometric method. The analysis showed that the water solution of the cefadroxil exhibited the highest peak abundance at 232 nm. The outcome of the study was matched with the previous literature and strongly supported our study. A report published by Shantier et al., evaluated the absorption maxima of cefadroxil in water and reported at 214 nm12.
Method validation analysis for quantitative evaluation of pharmaceutical drugs is one of the important approaches for their regulatory purpose. Several techniques have been associated with qualitative and quantitative evaluation, so far UV spectrophotometric analysis is still influencing the analytical techniques in the research area12. The method validation analysis was performed as per the described method in ICH guidelines. Linearity, LOD, LOQ, precision (%RSD), accuracy and robustness were determined for the developed and validated method. the outcome of the study showed the developed validated method was found linear, accurate, precise and robust at different concentrations of cefadroxil ranging from 05-30 µg/ml. The calibration equation and regression coefficient for developed method of gallic acid was found as y = 0.0225x + 0.0479 andR² = 0.9971, respectively. The average LOD and LOQ for the validated method were found as 0.583 ± 0.0012 and 1.767 ± 0.0027 at different concentrations. Precision was determined as intra-day precision and inter-day precision or relative standard deviation (%RSD). The results showed that intraday and inter day precision as % RSD 0.4293-2.2319 and 0.549-2.1409, respectively. Percentage drug recovery of the cefadroxil was determined as the accuracy of the developed and validated method by percentage spiking of the standard to the sample with 0, 50, 100, and 150%. The percentage of drug recovery was found ranging from 103.651-108.001%. The UV absorption spectrum and a calibration curve of cefadroxil are summarized in Figure 1.
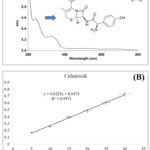 |
Figure 1: UV spectrum of cefadroxil (A) and calibration curve (B). |
Determination of dissolution rate of different tablets
Dissolution rate is one of the key factors not only for the development of effective pharmaceutical dosage forms but also to maintain the quality of the derived products 18. In the body, the drug dissolution rate depends on the solubility of active pharmaceutical ingredients and the particle size. It can be considered that lowering the particle size increases the dissolution rate of the dosage form especially tablets or capsules. By reducing the particle size (micronization and nanosizing) of both actives or excipients, the dissolution rate can be improved for the better efficacy of the drug 19. Considering the facts, the dissolution rate of different tablets of cefadroxil was determined using the standard protocol, successfully. the study was restricted to instrument conditions and time. Several samples were withdrawn during the experiment period to determine the dissolution rate of the drug based on the percentage of cefadroxil content found at different times of intervals. the outcome of the analysis was represented statistically using a Two-way ANOVA analysis followed by comparing the columns data. The significance level was considered in form of the p-value and the significant difference was determined at p<0.05. The outcome of the study showed that the highest percentage release of the drug was found 81.665 ± 2.354%, 79.685 ± 1.365%, 83.234 ± 1.336%, 79.234 ± 2.032% at 30 min. Furthermore, no significant difference was determined in the percentage release of the drug in the samples withdrawn after 25 min. The rate of dissolution of each tablet was found comparable. The results of the study have been summarized in Figure 2.
 |
Figure 2: Dissolution rate of different cefadroxil tablets. The data are represented as Mean ± SD (n=3) using Two-way ANOVA to compare all the pairs of columns. The significance level was considered as p<0.05. |
ADME/TOX analysis
ADME and toxicity prediction analysis of cefadroxil was done using Swiss ADME and Pro Tox tool, successfully. the study was conducted based on the prediction as mentioned in the standard guidelines of SWISS and a globally harmonized system of classification of labeling of chemicals. Parameters such as TPSA, consensus Log Po/w, ESOL Log S values, BBB permeant, GI absorption and log Kp (cm/s) (skin permeation) were predicted to determine the ADME, lipophilicity and the drug-likeness response of cefadroxil. TPSA act as a useful descriptor for the estimation of ADME properties such as concerning absorption and brain access 20. The TPSA value for cefadroxil was found as 167.49. The consensus log Po/w is anticipated by lipophilicity which represents the lipophilicity of anticipated molecules 21. The models accelerate the prediction accuracy for the physicochemical properties through consensus log Po/w which was found as -0.67. A positive value for logP represents the lipophilicity of the molecule while a negative value represents the hydrophilicity of the compound. Similarly, a model provided by Potts and Guy represents the skin permeability coefficient (Kp) and interrelated it with the molecular size and lipophilicity of the molecule. The more negative log Kpvalue represents the less permeant of the drug to the skin 13. Our findings suggest that several metabolites possess high skin permeability, as the log Kp value for cefadroxil was found as -10.42. The blood-brain barrier (BBB) permeant affinity of the molecules mainly depends on two different factors such as consensus log Po/w and TPSA represent apparent polarity or lipophilicity. In case, If the molecule covers the egg-shaped yolk part, it means that the molecule exhibits high BBB permeation, while it remains within the range of the white that characterizes HIA absorption 13. The outcomes of our study suggest that cefadroxil has low permeant affinity as it was found outside in the boiled egg plot of ADME analysis. In toxicity analysis, it has been revealed that cefadroxil has LD50- 10000 mg/kg and falls under the Predicted Toxicity Class: 6. The ADME egg and radar plot has been summarized in Figure 3.
 |
Figure 3: SWISS ADME analysis of cefadroxil, figure (A) represent boiled egg plot of cefadroxil showing the non-lipophilic nature of the drug. Figure (B) represents the chemical structure of cefadroxil while figure. |
In-silico analysis for determination of anti-inflammatory activity of cefadroxil
In-silico analysis was performed to determine the effect of cefadroxil against the inflammatory cytokines namely TNF-α and IL-6. The analysis was conducted through the Auto dock tool and the binding affinity of cefadroxil with the targeted proteins was determined in form of conventional hydrogen bond interaction. Three different sites for each protein were selected for the ligand interaction. The outcome of the study showed that the interaction of cefadroxil with TNF-α was found significant at each site with major interaction with the protein. The results of the study have been depicted in Figure 4.
 |
Figure 4: In-silico docking analysis of cefadroxil against TNF-α and IL-6. 3d and 2d representations have been shown in the figures. |
The outcome of the study showed that the binging affinity of cefadroxil showed a prominent interaction with TNF-α at site 1 with the binding -6.8. meanwhile, cefadroxil showed the prominent interaction at two different sites (site 1 and site 2), the exhibited binding energy at both sites was found as -6.8. The outcomes of the study were matched with the reported article and found that cefadroxil exhibits a comparable binding affinity with TNF-α and IL-6 cytokines protein. A study reported by Kim et al., evaluated the binding affinity of majonoside-R2 and ginsenoside R2 with TNF-α and reported the comparable outcomes in form of binding energy found as -8.1 and -7.9, respectively 22. In the studies reported by Aher and Perera evaluated biological interaction of karaviloside VI, karaviloside VIII, momordicoside L, momordicoside A, etc against IL-6 and reported that each compound possessed strong interaction with IL-6 23, 24.
Network pharmacology analysis for anti-inflammatory analysis of cefadroxil
In network pharmacology study, out of 150 selected genes, only two genes namely SLC15A1 and SLC15A2 were found active with significant interaction with cefadroxil. It has been reported that these such proteins are accountable for the dietary protein digestion products by absorption and conservation in the intestine as well as the kidney, respectively. They have been reported that these peptides are responsible for preserving the brain neuropeptides homeostasis. Absorption as well as the disposition of several therapeutically active molecules such as angiotensin-converting enzyme inhibitors, amino cephalosporins, antiviral drugs, etc facilitated by these proteins. Inflammatory bowel disease (Ser117Asn SNP and upregulation of colonics) and Lead exposure (2 haplotypes related to elevated blood lead consignment in the children, especially males) are the major malfunctions associated with the up and down-regulation of SLC15A1 and SLC15A2 25. In a report published by Song et al., reported that the associate’s genes of SLC15 such as SLC15A3 expression confirms bacterial peritonitis and typical findings comprehend that SLC15A3 is delimited by various TLRs. Moreover, these proteins play an essential role in the regulation of inflammatory response mediated by TLR426. A study conducted by Wang et al., evaluated that membrane transporter PhT2 (SLC15A3) which is acknowledged as the transporter of lysosomal membrane and up-regulated by lipopolysaccharide (LPS) through NF-κB signaling pathway, hence plays an important role in the expression of inflammatory cytokines27. The results have been depicted in Figure 5.
 |
Figure 5: Network pharmacology analysis of cefadroxil, the figure represents the strong interaction of the drug with SLC15A1 and SLC15A2 genes. |
Conclusion
The present study concludes that the developed and validated method found linear, accurate, precise and robust against the wide concentration of cefadroxil which might help for qualitative and quantitative validation of cefadroxil in different targeted and non-targeted samples. The dissolution rate of each tablet was found comparable while in-silico docking analysis and network pharmacology analysis showed a significant anti-inflammatory potential of cefadroxil via regulation of genes such as TNF-α, IL-6, SLC15A1 and SLC15A2. However, due to its bioavailability barriers, further experimental strategies are necessary to re-purpose the therapeutic application of cefadroxil as a potent anti-inflammatory agent.
Acknowledgment
The authors would like to thank Sanskar College of Pharmacy & Research (SCPR), Sanskar Educational Group, Ghaziabad, Uttar Pradesh, India-201302, for providing facilities to complete the research project and tech writeup for drafting the original manuscript.
Conflict of interest
The authors declare no conflict of interest.
Funding Sources
There is no funding Source.
References
- Six, R.; Cleaver, D.M.; Lindeman, C.J.; Cherni, J.; Chesebrough, R.; Papp, G.; Skogerboe, T.L.; Weigel, D.J.; Boucher, J.F.; Stegemann, M.R. J. Am. Vet. Med. Assoc.,2009, 234, 81-7.
CrossRef - H. Moglad, E.; Fatima, F.; Muqtader A, M.; Devanathad, V.; Khalid Anw, M.; F. Aldawsa, M. Int. J. Pharmacol.,2020, 16, 298-309.
CrossRef - Buck, R.E.; Price, K.E. Antimicrob. Agents Chemother.,1977, 11, 324-30.
CrossRef - Kesarla, R.S.; Vora, P.A.; Sridhar, B.K.; Patel, G.; Omri, A. Drug Dev. Ind. Pharm.,2015, 41, 1499-511.
CrossRef - Fylaktakidou, K.; Hadjipavlou-Litina, D.; Litinas, K.; Nicolaides, D. Curr. Pharm. Des.,2005, 10, 3813–3833.
CrossRef - Fan, X.H.; Cheng, Y.Y.; Ye, Z.L.; Lin, R.C.; Qian, Z.Z. Anal. Chim. Acta.,2006, 555, 217-24.
CrossRef - Harinantenaina, L.; Tanaka, M.; Takaoka, S.; Oda, M.; Mogami, O.; Uchida, M.; Asakawa, Y. Chem. Pharm. Bull.,2006, 54, 1017-21.
CrossRef - Yan, C.; Liu, H.; Lin, L. Biomed. Chromatogr.,2013, 27, 228-32.
CrossRef - Khan, S.B.; Alamry, K.A.; Alyahyawi, N.A.; Asiri, A.M. Int. J. Nanomedicine.,2018, 13, 3203.
CrossRef - Govindarajan, R.; Tejas, V.; Pushpangadan, P. J. AOAC Int.,2019, 102, 986-92.
CrossRef - Li, Y.; Wang, L.; Xu, B.; Zhao, L.; Li, L.; Xu, K.; Tang, A.; Zhou, S.; Song, L.; Zhang, X.; Zhan, H. J. Diabetes Res.,2021, 8891093.
CrossRef - Shantier, S.W.; Gadkariem, E.A.; Ibrahim, K.E.; El-Obeid, H.A. E-Journal Chem.,2011, 8, 1314-22.
CrossRef - Daina, A.; Zoete, V. Chem. Med. Chem.,2016, 11, 1117-21.
CrossRef - Rahman, N.; Muhammad, I.; Nayab, G.E.; Khan, H.; Aschner, M.; Filosa, R.; Daglia, M. Biomolecules., 2019, 9, 544.
CrossRef - Eswaramoorthy, R.; Hailekiros, H.; Kedir, F.; Endale, M. Adv. Appl. Bioinforma. Chem.,2021, 14, 13.
CrossRef - Islam, M.R.; Zaman, A.; Jahan, I.; Chakravorty, R.; Chakraborty, S. J. Young Pharm.,2013, 5, 173-9.
CrossRef - Yi, F.; Li, L.; Xu, L. jia, Meng, H.; Dong, Y. mao, Liu, H. bo, Xiao, P. gen.Chinese Med. (United Kingdom). 2018, 13, 1-20.
- Molavi, F.; Hamishehkar, H.; Nokhodchi, A. Adv. Pharm. Bull.,2020, 10, 656.
CrossRef - Dizaj, S.M.; Vazifehasl, Z.; Salatin, S.; Adibkia, K.; Javadzadeh, Y. Res. Pharm. Sci.,2015, 10, 95.
- Daina, A.; Michielin, O.; Zoete, V. Sci. Rep. 2017, 7, 1-3.
CrossRef - Mannhold, R.; Poda, G.I.; Ostermann, C.; Tetko, I. V. J. Pharm. Sci.,2009, 98, 861-93.
CrossRef - Kim, O.T.P.; Le, M.D.; Trinh, H.X.; Nong, H. V. Biophys. Physicobiology.,2016, 13, 173-80.
CrossRef - Aher, A.; Udhwani, T.; Khandelwal, R.; Limaye, A.; Hussain, T.; Nayarisseri, A.; Singh, S.K. Curr. Comput. Aided.Drug Des.,2019, 16,641-53.
CrossRef - Perera, W.H.; Shivanagoudra, S.R.; Pérez, J.L.; Kim, D.M.; Sun, Y.; Jayaprakasha, G.K.; Patil, B.S. Molecules.,2021, 26, 1038.
CrossRef - Smith, D.E.; Clémençon, B.; Hediger, M.A. Mol. Aspects Med.,2013, 34, 323-36.
CrossRef - Song, F.; Yi, Y.; Li, C.; Hu, Y.; Wang, J.; Smith, D.E.; Jiang, H. Cell Death Dis.,2018, 9, 1-5.
CrossRef - Wang, Y.; Sun, D.; Song, F.; Hu, Y.; Smith, D.E.; Jiang, H. Mol. Pharm.,2014, 11, 1880-8.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.










