Anthracene and 1,8-Napthalimide Aminothiazole Hybrids: Synthesis, Antimicrobial Activity and Molecular Docking Studies
Rambabu Palabindela , Prabhakar Myadaravenia, Devendar Banothu
, Prabhakar Myadaravenia, Devendar Banothu , Rajashekar Korra
, Rajashekar Korra , Himabindu Mekala
, Himabindu Mekala and Mamatha Kasula
and Mamatha Kasula
Department of Chemistry, Kakatiya University, Warangal-506009, Telangana India.
Corresponding Author E-mail: mamathakasula2016@gmail.com
DOI : http://dx.doi.org/10.13005/ojc/380117
Article Received on : 27-Dec-2021
Article Accepted on : 28-Jan-2022
Article Published : 27 Jan 2022
Reviewed by: Dr.G.Dayana Jeyaleela
Second Review by: Dr. P. S. Kumar
Final Approval by: Dr. Nenad Ignjatovic
With our efforts to develop potent antimicrobial agents, a series of anthracene and 1,8-napthalimide aminothiazole analogues were synthesized and characterized by using fundamental spectral analysis. All of the synthesized hybrids were screened for their anti-microbial activities against the bacterial strains E. coli, B. subtillis, and S. aureus, and the fungal strains A. niger and C. albicans. Among the compounds investigated, compounds 4c, 4d, and 6c exhibited the most potent antimicrobial activity. Fluconazole and Norfloxacin were used as standard drugs for antifungal and antibacterial activity. The molecular docking investigation revealed that the compounds 4c, 4d and 6c displayed the lowest binding energy values within the promoter regions of the PDB ID (1JIJ, 4WMZ). The in vitro antimicrobial activity results are well corroborated with the molecular docking results.
KEYWORDS:Anthracene; Aminothiazoles; Antimicrobial activity; Docking studies; 1,8-Napthalimide
Download this article as:| Copy the following to cite this article: Palabindela R, Myadaravenia P, Banothu D, Korra R, Mekala H, Kasula M. Anthracene and 1,8-Napthalimide Aminothiazole Hybrids: Synthesis, Antimicrobial Activity and Molecular Docking Studies. Orient J Chem 2022;38(1). |
| Copy the following to cite this URL: Palabindela R, Myadaravenia P, Banothu D, Korra R, Mekala H, Kasula M. Anthracene and 1,8-Napthalimide Aminothiazole Hybrids: Synthesis, Antimicrobial Activity and Molecular Docking Studies. Orient J Chem 2022;38(1). Available from: https://bit.ly/3Iyps9A |
Introduction
In current history, epidemiological studies have established that infections produced by pathogenic bacteria and fungi have a considerable negative impact on human health. Drug resistance was found to be on the increase, posing a threat to the efficacy of antimicrobial therapy, according to large-scale surveillance. The rise of multidrug-resistant pathogenic microorganisms has prompted researchers to look into new drug treatments.1 As a result, research is increasingly focused on developing new antimicrobial agents that increase the bioactivity of existing medications while simultaneously targeting other targets to address resistance.2 Heterocyclic naphthalimides have developed as a promising molecular frameworks, because of their wide spectrum of medical applications, such as anti-inflammatory, anti-cancer, antiviral, antibacterial, antidepressant, and antifungal agents.3,4 Thiazoles have been used in the development of drugs for HIV infections,5 allergies,6 pain,7 inflammation,8 hypertension,9 schizophrenia,10 bacterial infections11 and hypnotics,12 as well as new inhibitors of fibrinogen receptor antagonists and bacterial DNA gyrase B10 with antithrombotic activity.13 Nowadays a variety of commercial medicines used also contain the thiazole moiety, which includes epothilones, sulfathiazole, dasatinib, and tiazofurins.
Keeping the attributes mentioned above and in continuation of our research work in designing new bioactive heterocyclic frameworks, we developed anthracene and 1,8-napthalimide based aminothiazole hybrids as potential bioactive compounds and screened for their antimicrobial activity. Further, the molecular docking study was performed by using topoisomerase II and lanosterol alpha demethylase inhibitor. In this study, the molecular docking results of the compounds were correlated with their antimicrobial activity results.
Experimental Section
Material and Method
All the solvents and reagents used for the synthesis were of analytical grade and used directly. The completion of the reaction was monitored by using RANKEM silica gel G and E-Merck precoated TLC plates and visualization was done by using a UV chamber. The melting points of the newly synthesized compounds were determined in open capillary tubes and incorporated accordingly. 1H NMR spectra were recorded On a Bruker 400 MHz spectrometer. 13C NMR spectra were recorded on a Bruker 100 MHz spectrometer. The mass spectra were recorded using the ESI–Mass spectrum.
Synthesis
Synthesis of anthracene aminothiazole derivatives (4a-e)
A mixture of 9-Anthraldehyde (1 mmol, 206 mg), thiosemicarbazide (1 mmol, 91 mg), and α-bromoketones (3a-e, 1 mmol) was taken in pure ethanol (10 mL) and a few drops of ACOH were added and the mixture was stirred for 3-5 hrs at 60-70 ºC. TLC was used to observe the reaction progress. After the completion of reaction, the mixture was filtered, washed with fresh water and EtOH to produce the final compounds (4a-e) shown in Scheme 1.
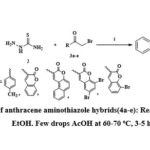 |
Scheme 1: Synthesis of anthracene aminothiazole hybrids(4a-e): Reagents and conditions: EtOH. Few drops AcOH at 60-70 oC, 3-5 h. |
Synthesis of naphthalimide aminothiazole derivatives (6a-e)
A mixture of 1,8-Naphthalic anhydride (5, 1 mmol), thiosemicarbazide (2, 1 mmol), and 3-α-bromoketones (3a-e, 1 mmol) was taken in 10 ml of DMF solvent, heated at 100 ºC for 3–4 hrs. The mixture was filtered and dried under vacuum to produce the final compounds (6a-e) displayed in Scheme 2.
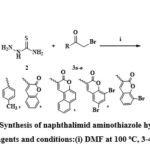 |
Scheme 2: Synthesis of naphthalimid aminothiazole hybrids(6a-e): Reagents and conditions:(i) DMF at 100 oC, 3-4 hrs. |
2-(2- (anthracen-9-ylmethylene) hydrazinyl)-4-(p-tolyl)thiazole (4a)
Yellow solid; mp: 258-260 ºC; proton-NMR values: δ 12.12 (N-NH), 9.20 (CH=N), 8.48-7.30(aromatic protons), 7.32 (thiazole C4-proton), 2.43 (-CH3); 13Carbon-NMR:δ 169.53, 152.35, 143.98, 138.54, 134.28, 130.39, 128.54, 127.98, 127.71, 127.16, 126.29, 110.06, 22.12; MS (C25H19N3S) m/z: 394.1215 [M + H]+.
3-(2-(2-(anthracen-9-ylmethylene) hydrazinyl)thiazol-4-yl) -2H-chromen-2-one (4b)
Yellow-solid; mp: 302-304 ºC; proton-NMR values: δ 12.10 (=N-NH), 8.89 (-CH=N), 8.53-7.36(aromatic protons) 7.26 (thiazole C4-proton), 7.23 – 7.18 (Aromatic-proton); 13Carbon-NMRvalues: δ 166.23, 159.83, 155.13, 153.58, 139.07, 132.87, 130.51, 130.28, 129.86, 129.56, 128.60, 127.94, 127.20, 126.32, 125.01, 121.15, 120.60, 117.14, 109.92; MS (C27H17N3O2S) m/z 448.1334 [M + H]+.
2-(2-(2-(anthracen-9-ylmethylene) hydrazinyl)thiazol-4-yl)-3H-benzo[f]chromen-3-one (4c)
Yellow solid; mp: 276-278 ºC;proton-NMR values: δ 12.11 (-N=NH), 9.08 (-CH=N), 8.59-7.39 (aromatic protons), 7.35 (thiazole C4-Proton);13Carbon-NMRvalues: δ 168.19, 160.23, 154.54, 152.55, 139.12, 131.06, 130.36, 128.96, 128.03, 127.21, 126.25, 124.80, 122.39, 121.68, 118.51, 117.08, 109.94; MS (C31H19N3O2S) m/z 498.1154 [M + H]+.
3-(2-(2-(anthracen-9-ylmethylene) hydrazinyl)thiazol-4-yl)-6,8-dibromo -2H-chromen-2-one (4d)
Orange-solid; mp: 312-314 ºC; proton-NMR values: δ 12.08 (-N=NH), 9.12 (, -CH=N), 8.62 – 7.41 (aromatic protons), 7.26 (thiazole C4-Proton); 13Carbon-NMRvalues: δ 168.23, 159.24, 152.36, 151.34, 139.11, 137.54, 131.25, 130.82, 130.42, 129.38, 128.68, 128.06, 127.16, 126.39, 121.89, 121.10, 119.57, 113.72, 108.99; MS (C27H15Br2N3O2S ) m/z 604.5316 [M + 2]+.
3-(2-(2-(anthracen-9-ylmethylene)hydrazinyl) thiazol-4-yl)-8-bromo-2H -chromen-2-one (4e)
orange-solid; mp: 318-320 ºC; proton-NMR values:δ 12.10 (-N=NH), 9.06 (-CH=N), 8.51-7.31 (aromatic protons), 7.22 (thiazole C4-Proton), 7.08 (Aromatic-proton); 13Carbon-NMRvalues: δ 168.53, 159.10, 153.35, 151.52, 139.35, 135.35, 130.68, 130.68, 129.32, 128.58, 128.12, 127.55, 126.58, 121.98, 120.64, 111.28, 109.12; MS (C27H16BrN3O2S) m/z 604.5316 [M + 2]+.
2-((4-(p-tolyl)thiazol-2-yl) amino)-1H-benzo[de] isoquinoline-1,3(2H)-dione (6a)
Pale yellow solid; mp: 228-230 ºC; proton-NMR values: δ 10.62 (-NH), 8.34-7.39 (aromatic protons), 7.21 (thiazole C4-Proton), 2.35 (-CH3); 13Carbon-NMRvalues: δ 168.74, 160.15, 143.89, 138.68, 134.56, 132.13, 131.64, 130.53, 129.99, 128.86, 127.82, 127.11, 126.34, 110.69, 22.78; MS (C22H15N3O2S) m/z 386.0912 [M +H ]+.
2-((4-(2-oxo-2H-chromen-3-yl) thiazol-2-yl)amino)-1H-benzo[de] isoquinoline-1,3(2H)-dione (6b)
Yellow solid;mp: 282-284 ºC;proton-NMR values: δ 10.84 (-NH), 8.38-7.53 (aromatic protons), 7.34 (thiazole C4-Proton), 7.20 (aromatic protons); 13Carbon-NMRvalues: δ 168.36, 161.23, 160.19 , 154.13, 139.24, 132.92, 132.06, 131.49, 129.92, 129.69, 128.81, 127.10, 126.39, 125.14, 121.20, 120.89, 117.20, 109.20; MS (C24H13N3O4S) m/z 440.3015 [M +H ]+.
2-((4-(3-oxo-3H-benzo[f] chromen-2-yl)thiazol-2-yl)amino)-1H-benzo[de] isoquinoline-1,3(2H)-dione (6c)
Yellow-solid; mp: 244-246 ºC;proton-NMR values:δ 10.85 (-NH), 8.36 (Ar-H), 8.25-7.32 (aromatic protons), 7.26 (thiazole C4-Proton); 13Carbon-NMRvalues: δ 168.39, 161.12, 160.02, 154.54, 139.36, 132.10, 131.46, 130.87, 130.15, 128.96, 127.09, 126.44, 124.62, 122.29, 121.61, 118.43, 116.83, 108.99; MS (C28H15N3O4S) m/z 490.0915 [M +H ]+.
2-((4-(6,8-dibromo-2-oxo-2H-chromen-3-yl) thiazol-2-yl)amino)-1H-benzo[de] isoquinoline-1,3(2H)-dione (6d)
Orange solid;mp:294-296 ºC;proton-NMR values:δ 10.79 (-NH), 8.36-7.53 (aromatic protons), 7.23 (thiazole C4-Proton); 13Carbon-NMRvalues: δ 168.29, 161.25, 159.76, 151.34, 139.26, 137.63, 131.98, 131.36, 129.99, 129.39, 128.84, 127.12, 126.39, 121.63, 121.35, 119.73, 113.67, 108.96; MS (C24H11Br2N3O4S) m/z 596.8832 [M +2]+.
2-((4-(8-bromo-2-oxo-2H-chromen-3-yl) thiazol-2-yl)amino)-1H-benzo[de] isoquinoline-1,3(2H)-dione (6e)
Orange solid; mp: 318-320 ºC;proton-NMR values: δ 10.79 (-NH), 8.38-7.43 (aromatic protons), 7.23 (thiazole C4-Proton); 13Carbon-NMRvalues: δ 168.34, 161.22, 160.04, 151.14, 139.21, 134.99, 131.98, 131.42, 130.16, 129.23, 128.79, 127.16, 126.39, 121.96, 120.42, 111.34, 109.05; MS (C24H12BrN3O4S): m/z 518.6302 [M + H]+.
Antimicrobial assay
The tube dilution method was used to test the antimicrobial properties of the developed anthracene and 1,8-nathptalimide aminothiazole analogues against Bacillus subtilis and Staphylococcus aureus (gram-positive bacteria) and Escherichia coli (gram-negative bacteria) and two fungal species, Candida albicans, and Aspergillus niger. The stock solutions were prepared for the synthesized hybrids (4a-e and 6a-e) as well as the standard drugs (fluconazole and norfloxacin) were taken in acetone to a conc. of 100 g/mL, and then serially tube diluted.14 Double strength sabouraud dextrose broth-I.P (antifungal) and strength nutrient broth-I.P (antibacterial) were used to dilution test.15 The samples have been incubated at 25±1 ºC for seven days (fungal), 37±1 ºC for 24 hours (bacteria), and findings were recorded in terms of MIC.
Molecular Docking
The molecular docking, a computer aided experimental approach was used to demonstrate the interacting mechanism of anthracene, and 1,8-napthalimide aminothiazole derivatives with receptor active sites. Chem Bio Design Ultra 12.0 (www.cambridgesoft.com) was used to draw all of the compounds 2D structures, and energy-minimized 3D structures. These were produced by using Gaussian 0916 and also by using a small basic set hf/3-21g*.17 Crystallography 3D structures of topoisomerase II for (bacterial), and lanosterol alpha demethylase for (fungal) proteins were retrieved from RSC PDB (www.rscb.org) PDB ID: 1JIJ, 4WMZ.18,19The proteins were cleaned and prepared for docking by using the UCSF-chimera 1.10.1 protein preparation wizard program, which removes any existing ligand and water molecules. For docking, such low-energy compounds and receptors are utilized. Auto-Dock 4.2 package suite and Auto-Dock Tools (ADT) version 1.5.6 were utilized for the molecular docking study.20 The docking procedure involves stiff protein receptors topoisomerase II and lanosterol alpha demethylase as well as flexible anthracene and 1,8-napthalimide aminothiazole analogues. ADT was utilized to remove the water molecules. Gasteiger charges were applied to verity atom, and integrated non-polar H atoms in protein structures. The distance between both the acceptor and donor atoms in an H-bond was determined to be 1.9 Å. The structure was saved in PDBQT format for further ADT research. For each analogue, ten docked conformations were created during docking. A genetic algorithm was utilized to calculate the energy. Rotatable bonds, gasteiger partial charges, non-polar hydrogen atoms and grid box with dimensions 72× 72 × 72 Å3 were created on the topoisomerase II, lanosterol alpha demethylase protein receptor was allocated with aid of spacing (Angstrom): 0.3750 Å and Auto Dock Tools 1.5.6.21 The maximum number of examinations and the population size were 2,500,000 and 150, respectively. Using Discovery studio 4.1.0, the output results were graphically evaluated.
Results and Discussion
Chemistry
All the synthesized compounds are stable at room temperature, the structural assignments were determined by using 1HNMR, 13C NMR, and ESI mass spectroscopic data. Spectral analysis for compound 4a reveals: 1H-NMR spectrum displayed hydrazinyl proton (–C=N-NH) at δ 12.12 ppm, imine proton (CH=N) at δ 9.20 ppm, and thiazole C4 proton at δ 7.32 ppm as singlet signals. The 13C NMR spectrum reveals major signals at δ 169.53 ppm for thiazole-C2 carbon, δ 143.98 ppm for imine carbon, δ 110.06 ppm for thiazole-C4 carbon, and δ 22.12 ppm for the methyl group. The ESI mass spectrum reveals [M + H]+ ion peak at m/z: 394.1215 which corresponds to the molecular weight of the compound 4a. As a result, the structure of compound 4a has been confirmed. Compound 6a reveals: Singlet signals were found in 1H NMR spectra for the –N-NH proton at 10.62 ppm and the thiazole-C4 proton at 7.21 ppm. The 13C NMR spectra have appeared signals at δ 168.74 ppm for thiazole-C2 carbon, δ 160.15 ppm for carbonyl carbon of 1,8-napthalimide, δ 110.69 ppm for thiazole-C4 carbon and δ 22.78 ppm for the methyl group. Similarly, the ESI mass spectrum of [M + H]+ ion peak at m/z: 386.0912 also further supports the formation of compound 6a.
Antimicrobial studies of compounds
Using the tube dilution method, researchers focus on one gram-negative bacteria, Escherichia coli, and two gram positive-bacteria, Bacillus subtilis and Staphylococcus aureus, and also on antifungal species Candida albicans, and Aspergillus niger. Fluconazole and Norfloxacin were used as standard references for fungi and bacteria, respectively. The MIC findings of the investigated compounds and reference medicines were reported in µM. The findings are summarized in Table 1. From the findings, compound 4c displayed excellent bacterial activity against the tested strains, exhibiting MIC values of 7.63 µM, 9.13 µM and 8.39 µM, respectively. The compounds 4d and 6c have displayed good activity against tested strains. The other compounds have moderate to poor activity against the three tested strains. These findings were compared to those obtained with the reference drug norfloxacin. The antifungal activity of compound 4d has exhibited promising activity against species A. niger and C. albicans, with MIC values of 7.31 µM and 8.64 µM, respectively. The compounds 4c and 6c have displayed good activity against the examined strains. The reaming compounds have moderate to poor activity compared with the reference drug fluconazole.
Table 1: Antimicrobial activity of the synthesized hybrids (4a-e and 6a-e)
|
Compounds |
Minimum inhibitory concentration (µM) |
||||
|
Bacterial |
Fungal |
||||
|
S. aureus |
B. subtilis |
E. coli |
A. niger |
C. albicans |
|
|
4a |
63.12 |
45.89 |
89.12 |
67.03 |
52.47 |
|
4b |
21.30 |
36.42 |
31.21 |
41.12 |
31.61 |
|
4c |
7.63 |
9.13 |
8.39 |
11.09 |
8.11 |
|
4d |
10.01 |
12.19 |
17.12 |
7.31 |
8.64 |
|
4e |
21.25 |
28.13 |
45.03 |
32.12 |
29.08 |
|
6a |
69.12 |
>100 |
48.74 |
>100 |
39.46 |
|
6b |
39.12 |
53.12 |
43.86 |
23.74 |
32.18 |
|
6c |
16.42 |
20.76 |
17.51 |
14.5 |
12.45 |
|
6d |
65.08 |
41.36 |
39.02 |
42.89 |
25.12 |
|
6e |
>100 |
75.23 |
29.67 |
36.45 |
63.69 |
|
Norfloxacin |
3.42 |
4.27 |
3.15 |
– |
– |
|
Fluconazole |
– |
– |
– |
3.85 |
4.56 |
Molecular docking
The binding mechanism of the synthesized anthracene and 1,8-napthalimide aminothiazole hybrids with their respective receptors was investigated by using molecular docking analysis. Most potent antibacterial hybrids (4c, 4d, and 6c), as well as a reference drug (norfloxacin), were studied using molecular docking as in receptor of topoisomerase-II (PDB ID: 1JIJ) as from the PDB file. The binding energies, interacting amino acids, and interacting groups were displayed in table-2. One of the most potent hybrids in these studies, i.e compound 4c is with a binding affinity of -12.56 kcal/mole, exhibited three H-bonding interactions with His 50, Asp 40, and Thr 75 amino acids, as well as with other hydrophobic interactions, π-π stacking with amino acid aromatic rings. The compounds 4d and 6c have the lowest binding energy values, with one hydrogen bond interaction with key amino acid Gly 193 by 4d and two hydrogen bond interactions with Asp 40 and Gly 193 key amino acid by 6c, as well as remaining hydrophobic interactions with aromatic rings of key amino acids. The best structural conformations of 4c, 4d, and 6c with receptor-binding pockets are shown in Figures 1.
 |
Figure 1: The best docking poses of the lead compounds (4c, 4d and 6c) with topoisomerase II (PDB ID: 1JIJ) |
The most potent antifungal scaffolds (4c, 4d, and 6c) of the reference drug (fluconazole) were compared to the lanosterol-α-demethylase binding poses from the PDB file 4WMZ. The binding energies, interacting amino acids, and interacting groups were displayed in table-3. The compounds 4c, 4d and 6c exhibited the lowest binding energies of -13.23 kcal/mol, -13.28 kcal/mol, and -12.93 kcal/mol. The molecule 4c has one H-bond interaction with the O atom of the carbonyl functional group on the coumarin moiety with Gln 479 amino acid residue, and the remaining interactions are all hydrophobic through amino acids. The compound 4d and 6c are with also lowest bonding energy values with two hydrogen-bonding interactions with amino acids His 468 by N atom of –NH group, and Ile 47 by N atom of thiazole ring by 4d and one H-bond interaction with key amino acid Gly 315 by O atom of carbonyl functional group on the naphthalimide ring by 6c and the remaining all the other hydrophobic interactions with amino acids. The best structural conformations of 4c, 4d, and 6c with receptor-binding pockets are shown in Figure 2.
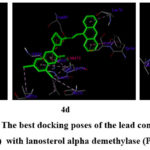 |
Figure 2: The best docking poses of the lead compounds (4c, 4d and 6c) with lanosterol alpha demethylase (PDB: 4WMZ) |
Table 2: (A) Molecular docking findings of most potent (antibacterial) analogues and standard drugs
|
Compounds |
Binding energy (kcal/mol) |
Interacting amino acids |
Interacting groups |
|
4c |
-12.56 |
His 50 |
N of –N=CH group |
|
Asp 40 |
N of thiazole ring |
||
|
Thr 75 |
O of carbonyl group on coumarin ring |
||
|
4d |
-11.77 |
Gly 193 |
O of carbonyl group on coumarin ring |
|
6c |
-11.71 |
Asp 40 |
O of carbonyl group on naphthalimide ring |
|
Gly 193 |
O of carbonyl group on coumarin ring |
||
|
Norfloxacin |
-6.66 |
Gln190 |
N of piperazin ring |
|
Lys 84 |
O of carbonyl group on carboxylic acid |
||
|
Arg 88, Asp 40 |
O of OH group on carboxylic acid |
||
|
Asp 80 |
O of OH group |
Table 2: (B) Molecular docking findings of most potent (fungal) analogues and standard drugs
|
Compounds |
Binding energy (kcal/mol) |
Interacting amino acids |
Interacting groups |
|
4a |
-13.23 |
Gln 479 |
O of carbonyl group on coumarin |
|
4c |
-13.28 |
His 468 |
N of –NH group |
|
Ile 47 |
N of thiazole ring |
||
|
6c |
-12.93 |
Gly 315 |
O of carbonyl group on naphthalimide ring |
|
Fluconazole |
-5.74 |
Ile 471 |
N of triazole ring |
|
Arg 385 |
Another N of triazole ring |
||
|
His 468 |
O of OH group |
Conclusion
In this study, anthracene and 1,8-napthalimide aminothiazole hybrids (4a-e and 6a-e) were synthesized, characterized and evaluated for antimicrobial activity and molecular docking studies were also performed. Among the synthesized analogues, 4c, 4d and 6c showed significant antimicrobial efficacy towards the bacterial and fungal strains. The molecular docking analysis of the potent analogues displayed the lowest binding energies. The in vitro antibacterial potency findings are well supported by the molecular docking results. Overall, current research shows that anthracene and 1,8-napthalimide aminothiazoles such as 4c, 4d and 6c have the potential for development as lead compounds, so further structural modifications could lead to promising new antimicrobial treatments.
Acknowledgement
The authors thank CSIR – UGC New Delhi for financial support and also thank the determent of chemistry Kakatiya University for providing research facilities.
Conflict of interest
The authors declare that they have no known competing financial interests
Funding Sources
There are no funding source.
References
- Zhang, H. J.; Qin, X.; Liu, K.; Zhu, D. D.; Wang, X. M, Zhu, D. D. Bioorg. Med. Chem. 2011, 19, 5708–5715.
CrossRef - Muhammad, Y. A.; Narang, R.; Nayak, S. K.; Singh, S. K. J Chem Pharm Res. 2016, 8(3), 930–937.
- Kamal, A.; Bolla, N. R.; Srikanth, P.S.; Srivastava, A. K. Expert Opin. Ther. Patents. 2013, 23, 299-317.
CrossRef - Zhang, Y. Y.; Zhou, C. H. Bioorg. & Med. Chem. Lett. 2011, 21 4349-4352.
CrossRef - Bell, F.W.; Cantrell, A. S.; Hoegberg, M.; Jaskunas, S. R.; Johansson, N. G.; Jordan, C. L.; Kinnick, M. D.; Lind, P.; Morin Jr, J. M. J. Med. Chem. 1995, 38(25), 4929-4936.
CrossRef - Haragave, K. D.; Hess, F. K.; & Oliver, J. T. J. Med. Chem. 1983, 26(8), 1158–1163.
CrossRef - Carter, J. S.; Kramer, S.; Talley, J. J.; Penning, T.; Collins, P.; Graneto, M. J.; Seibert, K.; Koboldt, C. M.; Masferrer, J.; Zweifel, B. Bioorg. & Med. Chem. Let. 1999, 9(8), 1171-1174.
CrossRef - Sharma, R. N.; Xavier, F. P.; Vasu, K. K.; Chaturvedi, S. C.; & Pancholi, S. S. J. Enzyme Inhib. Med. Chem. 2009, 24, 890–897.
CrossRef - Patt, W. C.; Hamilton, H. W.; Taylor, M. D.; Ryan, M. J.; Taylor Jr, D. G.; Connolly, C.J.; Doherty, A. M.; Klutchko, S. R.; Sircar, I. J. Med. Chem., 1992, 35(14), 2562-2572.
CrossRef - Jaen, J. C.; Wise, L. D.; Caprathe, B. W.; Tecle, H.; Bergmeier, S.; Humblet, C.C.; Heffner, T. G.; Meltzer, L. T.; Pugsley, T. A. J. Med. Chem., 1990, 33(1), 311-317.
CrossRef - Tsuji, K.; Ishikawa, H. Bioorg. Med. Chem. Lett. 1994, 4, 1601–1606.
CrossRef - Ergenç, N.; Çapan, G.; Günay, N. S.; Özkirimli, S.; Güngör, M.; Özbey, S.; Kendi, E. Inter. J. Pharm. Med. Chem. 1999, 332(10), 343-347.
CrossRef - Cappuccino, J.G.; Sherman, N. Addision Wesley Longman. Inc. Sydney, Australia. 1999, 477.
- Kumar, S.; Kaushik, A.; Narasimhan, B.; Shah, S. A. A.; Lim, S. M.; Ramasamy, K.; Mani, V. BMC Chem. 2019, 13(1), 1-17.
CrossRef - Pietro, W. J.; Francl, M. M; Hehre, W. J.; Defrees, D. J.; Pople, J. A.; Binkley, J. S. J. Am. Chem. Soc. 1982, 104, 5039-5048.
CrossRef - Frisch, M. J.; Trucks, G. W.; Schlegel, H. B. Gaussian 09, Revision B.01 Gaussian, Inc., Wallingford CT 2009.
- Qiu, X.; Janson, C. A.; Smith, W. W.; Green, S. M.; McDevitt, P.; Johanson, K.; Carter, P.; Hibbs, M.; Lewis, C.; Chalker, A.; Fosberry, A. Protein Science. 2001, 10(10), 2008-2016.
CrossRef - Sagatova, A. A.; Keniya, M. V.; Wilson, R. K.; Monk, B. C.; Tyndall, J. D. Antimicrob. Agents chemother. 2015, 59(8), 4982-4989.
CrossRef - http://autodock.scripps.edu/resources/references
- Zhang, J.; Zhao, J.; Wang, L.; Liu, J.; Ren, D.; Ma, Y. Tetrahedron. 2016, 72, 936-943.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.









