Antibacterial Activity and DNA Binding Properties of Bivalent Metal Complexes of Cuminaldehyde Acetoylhydrazone
Y.B. Nagamani2 , K. Hussain Reddy1*
, K. Hussain Reddy1* , K. Srinivasulu1
, K. Srinivasulu1 , D. Dhanalakshmi1
, D. Dhanalakshmi1 and K. Anuja1
and K. Anuja1
1Department of Chemistry, Sri Krishnadevaraya University, Ananthapuramu- 515 003, India.
2Department of Chemistry, Govt. Degree College (W), Hindupur, - 515201, India.
Corresponding Author E-mail: khussainreddy@yahoo.co.in
DOI : http://dx.doi.org/10.13005/ojc/370516
Article Received on : 21-08-2021
Article Accepted on : /pdf/vol37no5/OJC_Vol37_No5_p_21-08-2021.pdf
Article Published : 28 Sep 2021
Reviewed by: Dr. B Shivakumar

Second Review by: Dr. Basim Hatim

Final Approval by: Dr. Abdulwahab Omri
Metallo-hydrazones having the formula [M(IBAH)2] (where, M = Ni(II), Cu(II) and Zn(II); IBAH = p-Isopropylbenzaldehyde acetoylhydrazone) are prepared and confirmed on the basis of physico-chemical and spectral analyses. Conductivity data revealed that the complexes are non-electrolytes. Metal-DNA interactions are investigated using absorption spectrophotometry. Binding constant (Kb) data revealed that the copper complex interact DNA more strongly than other complexes. Antibacterial activity studies indicated higher activity for complexes than the metal free hydrazone ligand. The copper compound displays higher activity. DNA binding constants are correlated with the activity of metal compounds in this article.
KEYWORDS:Antibacterial activity; DNA binding constants; New Metallo-hydrazones; Spectral characterization
Download this article as:| Copy the following to cite this article: Nagamani Y. B, Reddy K. H, Srinivasulu K, Dhanalakshmi D, Anuja K. Antibacterial Activity and DNA Binding Properties of Bivalent Metal Complexes of Cuminaldehyde Acetoylhydrazone. Orient J Chem 2021;37(5). |
| Copy the following to cite this URL: Nagamani Y. B, Reddy K. H, Srinivasulu K, Dhanalakshmi D, Anuja K. Antibacterial Activity and DNA Binding Properties of Bivalent Metal Complexes of Cuminaldehyde Acetoylhydrazone. Orient J Chem 2021;37(5). Available from: https://bit.ly/2YR3tJl |
Introduction
Hydrazones exhibit a broad spectrum of biological activities such as antibacterial1, antitubercular1, antioxidant2, antiviral3 and anticancer4 activities. Hydrazones constitute an important class of compounds for new drug development5. Hydrazones derived from acetichydrazide are named as acetoylhydrazones. Compared to simple hydrazone Schiff bases, acyl and aroyl hydrazones have an extra donor site in >C=O. This presents a wide range of properties in them. The discovery that acetoylhydrazones show higher activity than benzoylhydrazones evoked considerable interest 6,7 to investigate metal complexes of former type of ligands.
Survey of literature revealed that a very few acetoylhydrazones are used as chromogenic reagents8 and in the study of transition 9-11 and lanthanide12, 13 metal complexes. Hydrazones derived from aldehydes contain hydrogen atom on azomethine carbon atom. It is reported14 that hydrazones having azomethine hydrogen atom shows higher activity. Hence it is of interest to investigate hydrazones derived from aldehydes rather than ketones.
Our customary foods contain carbonyl compounds15 showing beneficial effects to human health. The chemical name of cuminaldehyde is p– Isopropybenzaldehyde (IB). It is known to present in cumin, essential oils of eucalyptus, myrrh, cassia etc. The aromatic aldehyde, viz. para-Isopropybenzaldehyde (IB) is an important ingredient of cumin. It has medicinal properties. For example, it is known to inhibits the fibrillation of alpha-synuclein16 present in cumin. Hence it is of interest to use p– Isopropybenzaldehyde in the synthesis of new hydrazone ligand and its metal complexes.
Studies on Metal-DNA interactions are expected to predict biological activity of compounds. The compound which bind DNA strongly may find application in the drug development. Survey of literatures revealed that metal complexes with organic ligands showed higher biological activity than metal free organic compounds. Hence it is considered worthwhile to investigate novel hydrazone ligand originated from p– Isopropybenzaldehyde and its metal complexes. We have studied17-20 nucleic acid binding of various metal complexes in the past. to develop antimicrobial agents. To renew our interests, herein we communicate our results on spectral analysis and biological applications of bivalent metal complexes with p-Isopropylbenzaldehyde acetoylhydrazone (IBAH)
Experimental
Acetichydrazideand p-Isopropylbenzaldehyde were bought from Sigma Aldrich and utilized without further purification. Metal salts (ZnCl2, NiCl2.6H2O and CuCl2.2H2O) were of Merck AR quality. Solvents were distilled before use.
Synthesis of IBAH ligand
Round bottom flask(100 mL capacity) was charged with 20 mL of methanolic solution of acetic hydrazide (0.91g; 0.012 mol) and 20 mL of methanolic solution of p-Isopropylbenzaldehyde (5ml, 0.03 mol). Glacial acetic acid (few drops) was added as catalyst to the contents of flask. The reactants were heated on water bath for 2.5 hrs and cooled. The product was collected and treated repeatedly with hot H2O & dried. Methanol solvent was used in recrystallization of ligand. Yield:70.58%; M.P., 130-132oC. Molecular formula: C12H16N2O. Formula weight, 204.1. Synthesis of IBAH is depicted in Scheme 1.
 |
Scheme 1: Preparation of p-Isopropylbenzaldehyde acetoylhydrazone (IBAH) |
Preparation of metallo-hydrazones
The IBAH ligand (1.5 g; 0.007mol) was transferred to 100-mL beaker and dissolved in 20 mL ethanol. In another 100-mL beaker, CuCl2.2 H2O (0.007mol) was dissolved in 15 mL of ethanol. These two solutions were mixed in a clean R.B flask and refluxed for 2 hrs. On cooling, a green stained product was formed. The compound was amassed and washed with few drops 50% methanol and de-solvated in vacuum. Nickel(II) and zinc(II) complexes of IBAH were prepared similarly. Melting points and yields of compounds are delineated in Table 1.Particulars of equipment employed in the present study, deoxyribonucleic acid (DNA) binding and antibacterial activity experiments are given in our previous articles19, 20
Results and Discussion
The ligand (IBAH) is characterized on the basis of spectroscopic studies. FT-IR spectroscopy: 3264, 3082, 2945, 1648 and 1542 are designated to secondary amine (N-H), aromatic C- H, aliphatic C-H , >C=O and azomethine (>C=N) elongating vibrations correspondingly. 1H-NMR spectroscopy (in deuterated chloroform): δ 10.64 (s, 1H) 8.95 (s, 1H ) (7.29 – 7.75) (m, 4H), 2.32 (s, 3H) 1.271 (d, 6H) 2.97 (septet 1H) are respectively assigned to azomethine (CH=N-), >NH , Ar-H, CH3 of acetoyl, -CH3 protons of isopropyl and -CH methine protons of isopropyl group sequentially.. Mass spectrometry: Mass plot of IBAH (Fig. 1) shows a peak at m/z = 204.1 which coincides with formula (C12H16N2O) weight of IBAH.
 |
Figure 1: Mass spectrum of IBAH |
Based on spectral analysis the structure (Fig. 2) of IBAH ligand is proposed. Mass spectral fragmentation pattern of IBAH (Fig. 3) is shown below.
 |
Figure 2: Structure of IBAH ligand |
 |
Figure 3: Fragmentation pattern of IBAH ligand |
Structural characterization of metal complexes
Metal chlorides reacted with p-Isopropylbenzaldehyde acetoylhydrazone (IBAH) in basic medium under reflux conditions to produce corresponding metal complexes. The complexes are stable, non-hygroscopic, moderately soluble in CH3OH and C2H5OH but easily soluble in dimethylformamide(DMF) and dimethyl sulfoxide(DMSO). Color, yield, ESI-MS & conductivity data of Cu(II), Ni(II) and Zn(II) compounds are stated in Table 1. Molar conductivity outcome show that the coordination compounds are non-electrolytes21.
Table 1: Physicochemical and Analytical data* of complexes
|
Compound |
Colour (Yield, %) |
ESI-MS (F.W) |
Molar Conductivity@ |
|
Cu(IBAH)2 |
Dark green (74.36%) |
471.2 (469.5) |
12.75 |
|
Ni( IBAH)2 |
Parrot green (79.85%) |
464.8 (464.7) |
9.66 |
|
Zn( IBAH)2 |
Yellow (75.87%) |
468.3 (471.4) |
15.12 |
*Calculated values are given in parenthesis. @ Units, Ω-1cm2mol-1
Electronic spectroscopy
UV-Visible data of metal complexes and designation of peaks are summarized in Table 2. The electronic spectrum of Cu(II) complex shows(Fig. 4) peak at 14,792 cm-1 (676 nm) having shoulder peak at 16,447 cm-1 in the visible region. This band is assigned to 2Eg → 2T2g electron transfer in favour of octahedral geometry. The presence of shoulder peak at 16,447 cm-1 (608 nm) may be due to Jahn-Teller effect which is due to geometric distortion generally noticed for copper(II) octahedral compounds where the two axial bonds can be shorter or longer than those of the equatorial bonds.
 |
Figure 4: UV-Visible spectrum of Cu(IBAH)2 complex showing Jahn-Teller effect |
Table 2: UV-Visible spectral data of metal compounds with IBAH ligand
|
Complex |
Wavelength λmax (nm) (nm) |
Frequency (cm-1) |
Transition
|
|
Cu(IBAH)2 |
285 |
35087 |
π→π* |
|
608 676 |
16447 14792 |
d→d d→d |
|
|
Ni(IBAH)2 |
|
|
|
|
287 |
34843 |
π→π* |
|
|
611 |
16366 |
d→d |
|
|
989 |
10111 |
d→d |
Peaks are observed at 16,366 and 10,111 in the electronic spectrum of nickel complex. These peaks are related to 3A2g → 3T1g(F) and 3A2g → 3T1g(P) electron transfers respectively in support of octahedral geometry. A peak in higher energy region due to 3A2g → 3T2g transition is not observed due to the domination of π-→π* transition.
IR spectroscopy
The coordination sites ofIBAH ligand are uncovered by examining IR spectra of metal- free ligand and complexes. Spectral data are stated in Table 3. In the spectrum of the IBAH bands are observed due to νN-H, νC=C,νC=O andνC=N stretching modes. Both νN-H (3264 cm-1) and υC=O (1648 cm -1) bands of ligand are absent in the spectra of complexes due to enolization and subsequent deprotonation of ligand during complex formation (Scheme-2).
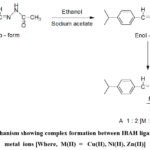 |
Scheme 2: Mechanism showing complex formation between IBAH ligand and bivalent metal ions [Where, M(II) = Cu(II), Ni(II), Zn(II)] |
Table 3: Infrared Spectral data of IBAH ligand and its complexes
|
IBAH |
Cu(IBAH)2 |
Ni(IBAH)2 |
Zn(IBAH)2 |
Assignment |
|
3264 |
– |
– |
– |
υN-H |
|
3082 |
3014 |
3027 |
3024 |
υC-H aromatic |
|
2945 |
2920 |
2915 |
2913 |
υC-H aliphatic |
|
1648 |
– |
– |
– |
υC=O |
|
1648 |
1615 |
1620 |
1612 |
υC=N |
|
1542 1465 |
1528 |
1522 |
1532 |
υC=C (aromatic) |
|
1378 |
1367 |
1318 |
1312 |
υC=C (aromatic) |
|
1264 |
1260 |
1242 |
1248 |
|
|
1185 |
1173 |
1178 |
1165 |
υC-O |
|
|
604 |
616 |
632 |
υM-O |
|
|
574 513 |
538 520 |
542 524 |
υM-N |
The bands of νC=O andνC=N areshifted to lower wave numbers indicating involvement of acetoyl >C=Oand azomethine (>C=N-) nitrogen groups in chelation. Spectral data suggest that the IBAH behaves as uninegative bidentate ligand in complexes. Bands in low energy regions, 604- 632 and 513-524 cm-1 are related to υM-O and υM-N vibrations respectively.
ESR spectroscopy
ESR spectrum of Cu complex in DMF at liquid nitrogen temperature (LNT) is depicted in Figure 5. Table-4 gives ESR data of copper(II) complex in solid state & in DMF solvent. The g║, g┴ and α2 and λ values of Cu complex suggest covalent nature of Metal- Ligand (M-L) bond and the location of unpaired electron in dx2 – y2 orbital. This finding indicates covalent nature 22 of metal- ligand (M-L) bond. The orbital reduction parameters (K║, K┴) reveal the occurrence of in-plane π- bonding in the complex. The axial symmetry parameter values of complex in solid and solution states are below 4.0. The values indicate absence of quid pro quo interaction.
 |
Figure 5: ESR spectrum of Cu(IBAH)2 at LNT |
Table 4: Spectral data† of [Cu (IBAH)2] complex in DMF at LNT
|
g|| |
g^ |
g avg |
G |
A||×10-5 |
A ^ ×10-5 |
K || |
K ^ |
λ |
α2 |
|
2.38 (2.20) |
2.07 (2.12) |
2.17 (2.15) |
5.64 (1.62) |
0.00262 |
0.00014 |
0.9970 |
1.091 |
520 |
0.3816 |
ESR data of complex at RT are given in brackets.
Based on analytical, molar conductivity, UV-Visible, FT-IR ESR spectroscopy studies a general structure (Fig. 6) is tentatively suggested.
 |
Figure 6: Proposed Structure for complexes in DMF medium |
Deoxyribonucleic acid Interactions
Interactivity of complexes with CT-DNA was examined by UV-visible spectroscopy. Spectra of copper complex are shown in Figure 7.
 |
Figure 7: UV-Visible spectra of copper complex with increasing amount of DNA. (Top most curve is spectrum of complex without DNA) |
Metal derivatives of IBAH showed strong peak due to M→L charge transfer transitions. The binding constant (Kb) is calculated using standard equation.23 DNA – binding constants of complexes are given in the Table-5. Metal complexes (except Cu(II) complex) showed considerable hypsochromic shift (Blue shift) (∆ λmax = 0.8 – 1.5 nm) with raising amounts of DNA. High binding constants (4.3- 17.2 x 106 M-1) of present complexes are suggestive 24-28 of intercalation mode of binding. The binding constant (Kb) order is Cu(IBAH)2 >Ni(IBAH)2 >Zn(IBAH)2. The observed trend indicates that Cu(IBAH)2 interacts DNA more firmly.
Table 5: UV-Visible Spectral Data Complexes with and without DNA
|
Complex |
λ max, nm (n) |
Δ λ |
H% |
Kb [M-1] X106 |
|
|
Free |
Bound |
||||
|
Cu(IBAH) 2 |
285.0 |
285.5 |
0.5 |
19.32 |
17.2 |
|
Ni(IBAH)2 |
286.0 |
284.5 |
-1.5 |
11.26 |
13.4 |
|
Zn(IBAH)2 |
242.2 |
241.4 |
-0.8 |
16.42 |
4.3 |
Antibacterial activity studies
The compounds under investigation are tested for antibacterial activity as outlined before5 by taking Gram-positive (Micrococcus luteus, Bacillus subtilis ) and Gram-negative (Escherichia coli, Pseudomonas aeruginosa) bacteria using agar well diffusion method against ciproflaxin as positive control. About 24 hrs old bacterial cultures are used to prepare bacterial lawns. Agar wells were prepared and injected with sample solutions prepared using DMF. The agar plates are incubated at 35oC for 24 hrs and inhibition zones are measured and expressed (Table 6) in millimeter units.
Table 6: Zone inhibition (in mm) data of IBAH ligand and its metal complexes
|
Compound |
Amount Taken (μg/μL) |
E.coli |
P.aeruginosa |
M.luteus |
B.substilis |
|
Ciprofloxacin |
5 |
11.68±0.05 |
10.24±0.01 |
9.78±0.02 |
10.36±0.04 |
|
IBAH |
100 |
1.57±0.19 |
1.56±0.11 |
1.49±0.37 |
1.03±0.43 |
|
200 |
1.82±0.35 |
2.48±0.01 |
2.73±0.25 |
2.87±0.04 |
|
|
300 |
2.64±0.44 |
2.65±0.28 |
2.68±0.08 |
2.29±0.22 |
|
|
Cu( IBAH) 2 |
100 |
6.45±0.25 |
3.42±0.39 |
4.09±0.32 |
3.93±0.16 |
|
200 |
7.05±0.36 |
5.46±0.05 |
5.92±0.42 |
5.84±0.38 |
|
|
300 |
6.31±0.08 |
6.29±0.03 |
6.38±0.14 |
5.68±0.24 |
|
|
Ni( IBAH)2 |
100 |
2.14±0.06 |
1.75±0.06 |
1.92±0.42 |
2.81±0.21 |
|
200 |
3.95±0.18 |
2.22±0.03 |
2.33±0.50 |
2.70±0.13 |
|
|
300 |
4.09±0.22 |
4.86±0.36 |
3.57±0.27 |
4.58±0.11 |
|
|
Zn( IBAH)2 |
100 |
2.02±0.17 |
3.08±0.23 |
2.73±0.04 |
1.89±0.19 |
|
200 |
4.94±0.01 |
4.33±0.18 |
4.52±0.41 |
2.51±0.28 |
|
|
300 |
5.73±0.23 |
6.14±0.31 |
3.43±0.29 |
4.73±0.35 |
Results in Table 6 indicate that the present coordination compounds show higher activity than metal-free IBAH ligand. Data are depicted in the form of bar graph (Fig. 8) which indicates that Cu(IBAH)2 complex shows more activity possibly due to its strong binding to DNA.
 |
Figure 8: Graphical representation of inhibition zones (in mm) of metal complexes |
Metal complexes displayed higher antibacterial activity28,29 than the metal free ligand. Increased activity of coordination compounds may be explained by using Tweedy’s chelation theory30 and Overtone’s concept31 . The ligand forms organic coat around the metal ion in the formation of complex. As a result, the complex becomes lipophilic. According to latter concept, the cell membrane allows the passage of complexes, as they are lipid-soluble. Hence, lipid solubility of compound is principal requirement for showing this action. On complex formation, the polarity of metal ion is significantly reduced due to delocalization of π-electrons. On entering into the cell, the complex releases metal ion , which inhibits enzymatic activity of microorganisms more effectively32.
Conclusion
A new organic ligand viz. p– Isopropylbenzaldehyde acetoylhydrazone (IBAH) and it’s metal complexes are synthesized and characterized for the first time. A general structures for the complexes are proposed based on analytical and electronic spectral data. High DNA binding constants suggest that the complexes bind DNA via intercalation. Complexes showed higher antibacterial activity. Among all compounds, the copper complex binds DNA more firmly and inhibits bacteria effectively. The Cu(IBAH)2 complex not only binds DNA strongly but also inhibits bacteria more effectively. The observation revealed a direct relationship between DNA binding constant and antibacterial activity. Thus, antibacterial activities of metal complexes are concomitant with their DNA binding constants.
Acknowledgement
K. H. Reddy is grateful to UGC, New Delhi for the award of BSR Faculty Fellowship.
Conflicts of interest
Regarding the publication of this article, the authors declare that there is no conflict of interests.
References
- Raja, A.S.; Agarwal, A.K.; Mahajan, N.; Pandeya, S.N.; Ananthan, S. Antibacterial and antitubercular activities of some diphenyl hydrazones and semicarbazones., Indian J Chem., 2010, 49B(10), 1384–1388.
- Rajitha, G.; Saideepa, N.; Praneetha, P. Synthesis and evaluation of N-(α-benzamidocinnamoyl) aryl hydrazone derivative for anti-inflammatory and antioxidant activities., Indian J Chem., 2011, 50B, 729–33.
CrossRef - Tian, B.; He, M.; Tang, S.; Hewlett, I.; Tan, Z.; Li, J. et al. Synthesis and antiviral activities of novel acylhydrazone derivatives targeting HIV-1 capsid protein., Bioorg Med Chem Lett., 2009, 19, 2162–2167.
CrossRef - Altýntop, M.D.; Özdemir, A.; Turan-Zitouni, G.; Ilgýn, S.; Atlý, Ö.; Ýþcan, G. et al. Synthesis and biological evaluation of some hydrazone derivatives as new anticandidal and anticancer agents., Eur J Med Chem., 2012, 58, 299–307.
CrossRef - Kasimbi, D.; Hussain Reddy, K.; Devanna, N. Synthesis, spectral characterization and antibacterial activity of functionalized hydrazones., Oriental J. Chem., 2019, 35, 557- 562.
CrossRef - Kasimbi, D.; Hussain Reddy, K.; Devanna, N. Synthesis, spectral characterization and antibacterial activity of copper(II) complexes functionalized hydrazones., Asian J. Chem., 2019., 31, 289-1293.
CrossRef - Renuka, M.; Hussain Reddy, K. Non-extrative spectrophotometric determination of copper in alloys and ores using 2,4-dihydroxyacetophenone acetoylhydrazones., Research Journal of Pharmaceutical, biological and chemical sciences., 2014, 5, 603-612.
- Nagakavitha, D.; Hussain Reddy, K. Synthesis, spectral characterization and DNA binding properties of dinuclear nickel(II) complexes with pyridine based hydrazones., International Journal of Pharma and Biosciences., 2014, 5, 294-304.
- Nagakavitha, D.; Hussain Reddy, K. Synthesis, spectral characterization and DNA binding properties of copper(II) complexes of functionalized hydrazones., Research Journal of Pharmaceutical, Biological and chemical sciences., 2014, 5, 1806-1815.
- Nagakavitha, D.; Hussain Reddy, K. Synthesis, spectral characterization and DNA binding properties of dinuclear copper(II) complexes with pyridine hydrazones., J. Indian Chem. Soc., 2015, 92, 71-77.
- Raja, K.; Suseelamma, A.; Hussain Reddy, K. Synthesis, X-ray crystal structure, DNA binding and nuclease activity of lanthanide(III) complexes of 2-benzoylpyridine acetoylhydeazone., Journal of Chemical Sciences., 2016, 128, 1265-1275.
CrossRef - Hussain Reddy, K.; Raja, K.; Suseelamma, A. Synthesis, Crystal structure, DNA binding and cleavage activity of butterfly-like 12-coordinate praseodymium (III) complex of 2-formypyridine acetoylhydeazone., Inorganic and Nanometal Chemistry., 2017, 47, 1398-1405.
CrossRef - Moussa, Z.; Al-Mamary, M.; Al-Juhani, S.; Ahmed, S.A. Preparation and biological assessment of some aromatic hydrazones derived from hydrazides of phenolic acids and aromatic aldehydes., Heliyon., 2020, 6, e05019.
CrossRef - Shahverdi, A.R.; Monsef-Esfahani, H.R.; Tavasoli, F.; Mirjani, R. Trans-cinnamaldehyde from Cinnamomum zeylanicum bark essential oil reduces the clindamycin resistance of Clostridium difficile in vitro., Journal of Food Science., 2007, 72, S055-S058.
CrossRef - Morshedi, D.; Aliakbari, F.; Tayaranian-Marvian, A.; Fassihi, A.; Pan-Montojo, F.; Pérez-Sánchez, H. Cuminaldehyde as the major component of cuminum cyminum, a natural aldehyde with inhibitory effect on alpha-synuclein fibrillation and cytotoxicity., Journal of Food Science., 2015, 80, H2336–H2345.
CrossRef - Moksharagni, B.; Hussain Reddy, K. A review on pharmaceutical applications of Lanthanide complexes with nicotinoyl and isonicotinoyl hydrazones., European Journal of Biomedical and Pharmaceutical Sciences., 2018, 58, 810 -817
- Chandrasekhar, S.; Hussain Reddy, K. DNA binding and nuclease activity of Structurally characterized copper(II) complex., International Journal of Pharmaceutical Sciences and Research., 2016, 7, 4204-4213.
- Moksharagni, B.; Rishitha, M.; Hussain Reddy, K. Synthesis, DNA binding properties and antibacterial activity of lanthanide complexes with 2-benzoylpyridine isonicotinoylhydrazone., Indian J Chem., 2017, 56, 232- 237.
- Moksharagni, B.; Hussain Reddy, K. Spectral characterization and DNA binding Properties of lanthanide(III) complexes with 2-acetylpyridine isonicotinoylhydrazone (APINH)., Bull Chem Soc Ethiopia., 2016, 30, 221-230.
CrossRef - Geary, W.J. The Use of Conductivity Measurements in Organic Solvents for the Characterization of Coordination Compounds., Coord Chem Rev., 1971, 7, 81-122.
CrossRef - Hussain Reddy, K.; Sambasiva Reddy, P.; Babu, P.R. Synthesis, spectral studies and Nuclease activity of mixed ligand copper (II) complexes of heteroaromatic semicarbazones/thiosemicarbazones and pyridine., Journal of Inorganic Biochemistry 1999, 77, 169-176.
- Wolfe, A.; Shimer, G.H.; Meehan, T. Polycyclic aromatic hydrocarbons physically intercalate into duplex regions of denatured DNA., Biochemistry., 1987, 26, 6392-6396.
- Krishnamoorthy, P.; Sathyadevi, P.; Cowley, A.H.; Butorac, R.R.; Dharmaraj, N. Evaluation of DNA binding, DNA cleavage, protein binding and in vitro cytotoxic activities of bivalent transition metal hydrazine complexes., Eur J Med. Chem., 2011, 46(8), 3376-3387.
CrossRef - Kumar, P.; Gorai, S.; Santra, M.K.; Mondal, B.; Manna, D. DNA binding, nuclease Activity and cytotoxicity studies of Cu(II)complexes of tridentate ligands., Dalton Trans., 2012, 41, 7573.
- Zhang, R.; Wu, X.; Yalowich, J.C.; Hasinoff, B.B. Design, synthesis, and biological evaluation of a novel series of bisintercalating DNA-binding piperazine-linked bisathrapyrazole compounds as anticancer agents., Bioorg. Med. Chem., 2011, 19, 7023- 7032.
- Arjmand, F.; Parveen, S.; Afzal, M.; Shahid, M.J. Synthesis, charecterization, biological studies (DNA binding, cleavage, antebacterial and topoisomerase I) and molecular docking of copper(II) benzimidazole complexes., Photochem Photobiol B Biology.,2012, 114, 15- 26.
CrossRef - Sirajuddin, M.; Ali, S.; Badshah, A. Drug-DNA interaction and their study by UV-Visible, fluorescence spectroscopies and cyclic voltammetry., Photochem. Photobiol. B Biology., 2013, 124, 1-19.
- Prasad, R.V.; Thakkar, N.V. Study of cobalt complexes as catalysts in the decomposition of hydrogen peroxide., J. Mol. Catal., 1994, 92, 9-20.
CrossRef - Belaid, S.; Landreau, A.; Djebbar, S.; Benali-Baitich, O.; Bouet, G.; Bouchara, J.P. Synthesis, characterization and antifungal activity of a series of manganese(II) and copper(II) complexes with ligands derived from reduced N,N1-O-phenylene bis(salicylideneimine)., J. Inorg. Biochem., 2008, 102, 63-69.
- Tweedy, B.G. Plant Extracts with Metal Ions as Potential Antimicrobial Agents., Phytopathology., 1964, 55, 910-918.
- Dharmaraj, N.; Viswanathamurthi, P.; Natarajan, K. Ruthenium(II) complexes containing bidentate schiff bases and their antifungal activity., Transition Met. Chem., 2001, 26, 105- 109.
- Neelakantan, M.A.; Esakkiammal, M.; Mariappan, S.S.; Dharmaraja, J.; Jeyakumar, T.Synthesis, Characterization and Biocidal Activities of Some Schiff Base Metal Complexes Indian J. Pharm Sci., 2010, 72, 216-222.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.









