Synthesis, Biological Valuation and Molecular Docking Analysis of New 5-Benzylidene Bis-Rhodanine Derivatives
Rajendran Kumar and Subban Ravi*
Department of Chemistry Karpagam Academy of higher Education, Coimbatore-21
Corresponding Author E-mail: ravisubban@rediffmail.com
DOI : http://dx.doi.org/10.13005/ojc/360609
Article Received on : 13-10-2020
Article Accepted on : 17-11-2020
Article Published : 30 Dec 2020
The synthesis of 5-benzylidene bis-rhodanine derivatives are reported from bis-rhodanine (III) and different aromatic aldehydes (IV) via Knoevenagel condensation reactions. All the Derivatives (V) and (Va-m) were deep-rooted by NMR spectroscopic techniques and elemental analysis. The antiproliferativestudy of the compounds on HeLa human cervical cancer cell line, K562 leukemic cell line and MDAMB231 breast cancer cell line were performed by MTT assay. Docking studies were carried out against the protein HPV 16E2 present in the HeLa cell line. It show good docking scores. The results indicate that the bis-rhodanine derivatives could serve as potential molecules for the development of new anticancer agents.
KEYWORDS:Breast Cancer Cell Line; Bis-Rhodanine; Docking Studies; Hela Cell Line; Leukemic Cell Line
Download this article as:| Copy the following to cite this article: Kumar R, Ravi S. Synthesis, Biological Valuation and Molecular Docking Analysis of New 5-Benzylidene Bis-Rhodanine Derivatives. Orient J Chem 2020;36(6). |
| Copy the following to cite this URL: Kumar R, Ravi S. Synthesis, Biological Valuation and Molecular Docking Analysis of New 5-Benzylidene Bis-Rhodanine Derivatives. Orient J Chem 2020;36(6). Available from: https://bit.ly/3kJ8gSh |
Introduction
Heterocyclic compounds are a major part of synthetic therapeutic chemistry. They offer a high level of structure variety and have demonstrated to be generally utilized as therapeutic agents. They are distributed widely as a natural product. Heterocyclic compounds have great potential and the most encouraging molecule for the discovery of new drugs. Ever since the introduction of Ciglitazone and Epalrestat for the treatment of diabetes, rhodanine has attracted medicinal chemists showing favourable biological properties.[1,2,3] The chemical derivatization of rhodanine gives rise to molecules with a broad range of therapeutic activities like antiviral[4,5], antifungal[6,7,8], antibacterial[9-16], antitumor[17], aldose reductase inhibitors[18-20] and anti-inflammatory activities. Rhodanine has also been reported as inhibitors of uridinediphosphate-N-acetyl/L-alanine ligase [21], hepatitis C virus protease inhibitors [22] and also inhibitors of cancer cell migration [23-26]. From the laboratory,it has been reported 5-benzylidene-3-ethyl rhodanine and 5-isopropylidene-3- ethyl rhodanine induced cytotoxicity in a time and concentration-dependent manner with an IC50 value of < 10 µ M [27,28]. Recently focus has been givenon the mixture of bis-heterocyclic compounds, which showed various biological activities [29-32]. As part of researcher’s growing interest in synthesizing bis-rhodanine [33].we describe herein an easy and inexpensive synthetic route for the synthesis of a set bis-rhodanine to find novel and more potent anticancer agent.
Experimental Procedure
General procedure for the synthesis of Bis-rhodanine(III)
1.2-diamino propane(0.01mol)was dissolved in water and while stirring then sodium Hydroxide(0.04mo.l) was added. The solution was cooled at 0ºC, the solution of carbon disulfide (0.02mol)was mixed into it and kept for stirring for about 4h.Then the water solution of sodiumchloroacetate(0.02mol) was mixed with it and stirred for 3h.Subsequently, the solution of HCl was added and refluxed for 2-3Hr. While cooling a precipitate appeared and then it was filtered and dried to the corresponding product(III)
3,3’-(propane-1,2-diyl)bis(2-thioxothiazolidin-4-one): (III)

Synthesis of 5-benzylidene bis-rhodaninederivatives(V)&(Va-m)
The mixture of bis-rhodanine(III) (0.002 mol)and substituted aromatic aldehyde(IV)and(IVa-n) (0.004 mol) were refluxed in glacial acetic acid(0.004mol) and sodium acetate (0.004 mol)for 4-6Hr. The resultant mixture on cooling yielded a precipitate which was filtered, washed, dried and recrystallised(ethanol)to afford the corresponding product(V)&(Va-m).










Anticancer Activity
Cell Line
The cervical cell line HeLa, leukemic cell line K562 and the cell line MDAMB231 for breast cancer were received from NCCS-National Center for Cell Science, Pune and grown.
MTT Assay
MTT assay was followed by the standard protocol according to Rajendran Kumar et al. The worth of IC50 was calculated with a nonlinear regression graph that strategized amid current inhibition of cells in one axis and concentration of logs oneother.
LDH Assay
LDH release assay liberates, Lactate dehydrogenase (LDH) is a pointer of membrane integrity and henceforth cell injury. Following treatment with compound Vf(3.12, 6.25, 12.5, 25, 50 and 100 µg/ml) onHeLa, K562, MDAMB231 cancer cells for 48 hours and it was conducted to measure the proclamation of LDH on the way to the media and measured using standard protocols (Moorthy et al. 2010). The intracellular LDH was determined by fast congelation and melting in liquid nitrogen after losing the cells. The proclamation of LDH was calculated at an absorption rate of 490 nm. The releasing proportionof LDH was measured, for example (media LDH activity)/ (Media LDH activity) X 100%. Outcomes are exposed as a proportion of LDH release that substrates switchstandards from the preserved ones.
FACS Analysis
Analysis of the FACS was performed to determine compound 1 had any effect on the development of cell cycles. After 48h of 50 and 100 µg/ml treatments, MDAMB231 cells stained with propidium iodide and endangered to FACS. The DMSO treated cells showed a regular pattern for the cell rotation. Cells were treated with compound Vf absorptions of 50 and 100 µg/ml and incubated for further 24 hrs. At room temperature, cells were gathered then centrifuged for 5 mins at 2000rpm. The supernatant was carefully removed, preserving the cell pellet. The cell pellet washed through re-dangling in 2mlof 1XPBS. With the same conditions, the washing was repeated for another time. The supernatant abandoned, with the pellet was retained. Cells set by resuspending in 300µl of Sheath fluid. Preceded by adding 1 ml of chilled 70% EtOHdropwise with constant gentle shaking, and adding another 1 ml again.
Docking studies
The mark protein nominated for the current research is HPV 16 E2 protein. The manufactured bis- rhodanine derivatives were practiced for molecular docking study.
Molecular Docking
The study of Docking under the default setting was done using Autodock with a binding pocket on the TAD. The result was analyzed using PYMOL (TM) by visualizing the protein-ligand and calculating parameters like hydrogen bonding amid protein and the ligand; thus, determining scores are tabulated.
Result and Discussion
While doing the literature survey to the best of our knowledge, there is only one report available for the synthesis of bis-rhodanine using diamines,Carbon disulfide, and dialkylacetylenedicarboxylates [32]. In the present work,diamines, Carbon disulfide, and chloroacetic acid for the synthesis of bis-rhodanine are used. To found the optimal reaction conditions for the integration of bis-rhodanine, it is decided to begin with the conventional method. In this method, one mole of diamine and two moles of carbon disulfide and chloroacetic acid are used. The dithioamideis formed initially by the reaction of carbondisulfide, and diamine serves as a nucleophile and reacts further with chloroacetic acid followed by cyclization yield bis-rhodanine (III). This method havebeen reported earlier [33], but the mechanism was not explained adequately. Now the device is shown in scheme 2.
 |
Scheme 1: Synthesis of Bis-rhodanine. |
 |
Scheme 2: Synthesis of 5-benzylidene Bis-rhodanine derivatives |
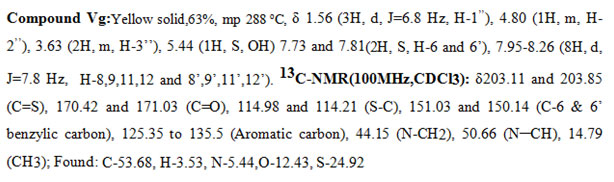
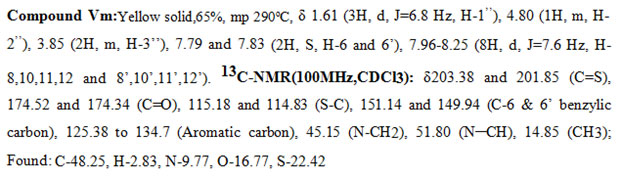
New derivatives were synthesized by Knoevenagel condensation of bis-rhodanine (III) with different substituted aldehydes (IV) in the presence of sodium acetate and acetic acid to yield the products (V) and (Va-m). The newly synthesized5-benzylidenebis-rhodanine derivatives were fully characterized by proton NMR, carbon NMR spectral data, and elemental analysis.
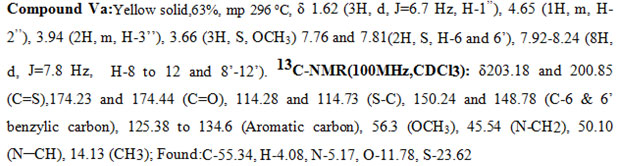
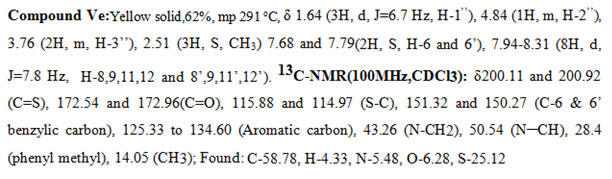
In the 1H-NMR spectra of a representative compound Ve, in the upfield region the N-methylene protons resonated at δ 3.76 (2H, m, H-3″), the methyl group appeared as a doublet at δ 1.64 ( 3H, J = 6.7 Hz), and the corresponding methine proton appeared as a multiplet at δ 4.8 (1H, m, H-2″). The two singlets at δ 7.68 (1H, s, H-6) and 7.79 (1H, s, H-6′) were attributed to the benzylic protons one on each side. The Phenyl protons appeared as a pair at δ 7.96 (4H, d, J = 7.8 Hz) and 8.15 (4H, d, J = 7.8 Hz). The CH3 group attached to the phenyl ring resonated at 2.51 (3H, S, CH3). The above data was complemented by its carbon NMR spectra. The pair of signals at δ200.11and200.92 and δ174.87 and 174.62 are due to thiocarbonyl (C=S) and carbonyl carbon (C=O) of the rhodanine ring system respectively. The pair of signals at δ 115.88 and 114.97 are due to the =C-S group of the rhodanine system. The CH3 carbon appeared at δ 14.05, and the CH2 and CH carbons attached to the nitrogen resonated at 50.54 and 43.26. The signals at δ151.32 and 150.27 are assigned to the benzylic carbons (C-6 & C-6′). The methyl carbon attached to the aromatic system appeared at δ28.4. The bunches of signalsbetween 125.33 to 134.60 are attributed to the phenyl carbon atoms. All the other messages of the compound (V) and (Va-m) coincided with signs of compound(Ve).
Mechanism
 |
Scheme 3: plausible mechanism for the formation of bis-rhodanine (III) and 5-benzylidene bis-rhodanine derivatives (V) and (Va-m) |
Cytotoxic Activity
The effect of compounds V and Va-m on the proliferation of HeLa, MDAMB231, and K562 cells were carried out using MTT assay. HeLa, MDAMB231, and K562 cells were added with 3.12, 6.25, 12.5, 25, 50, and 100µMof compounds V and Va-m and after 48hrs the assay was performed. IC50 values were calculated and presented in the Table1. The viability of the cells were affected at a moderate concentration in all the derivatives V and Va-m with HeLa, MDAMB231, and K562 cells, and was more pronounced in the case of the compound Vf against MDAMB231 breast cancer cell lines. The compound V without any substituents should be an IC50 value of 75.0, 85.4, and 72 µM concentration against HeLa, MDAMB231, and K562 cells.
The IC50 value of all the other compounds Va-m which has substituents in the aromatic system has varied substantially from the IC50 value of compound V against the three cell lines. This indicates that the substituents affect the proliferation activity against the three cell lines. If we divide these substituents into two groups like electron-withdrawing substituents (NO2,F, Cl and Br) and electron-pumping substituents (-OCH3, OH and CH3 ) and consider the IC50 value concerningHeLa cell lines, the activity in compounds with electron-withdrawing groups Vb, Vc, Vd, Vf showed lower IC50 costs than the compounds with electron-donating substituents Va, Ve and Vg. As an example, the IC50 value of compound Va with an electron-donating substituent – OCH3 is 81.2 µM.
In contrast, for the compound with electron-withdrawing substituent fluorine in Vf, the IC50 value is 54 µM. The same trend is followed in MDAMB231 and K562 cells too. Further the compounds are divided into two groups depending upon the position of the substituent placed in the aromatic ring-like para-substituted compounds Va to Vg and meta substituted compounds Vh, Vi, Vj, and Vm, the IC50 value of para-substituted compounds and relatively lower than the meta substituted compounds. The IC50 value of Vf with a para-substituted fluorine group has an IC50 value of 54 µM, whereas the corresponding compound with meta substituted fluorine group showed an IC50amount of 61 µM. Overall concerning all the three HeLa, MDAMB231, and K562 cell lines, the mixture Vf with a para-substituted fluorine atom performed well. It showed an IC50 value of 54.0, 32.0, and 58.0 against HeLa, MDAMB231, and K562 cell lines, respectively. So compound Vf was taken for further studies.
Table 1: Inhibition concentration(IC50) of compounds (V) and (Va-m) using HeLa cell line, leukemic cell line and breast cancer cell line by MTT assay and binding energy with 1DTO
|
S. No |
Compound |
R |
IC50,µM HeLa |
MDAMB231 |
K562 |
Binding energy, kcal/mol with 1DTO |
|
1 |
(V) |
H |
75.0 |
85.0 |
72.0 |
-8.6 |
|
2 |
(Va) |
p-OCH3 |
81.2 |
82.5 |
78.2 |
-9.5 |
|
3 |
(Vb) |
p-Cl |
77.0 |
78.0 |
79.0 |
-9.6 |
|
4 |
(Vc) |
p-Br |
55.5 |
65.2 |
60.5 |
-8.9 |
|
5 |
(Vd) |
p-NO2 |
66.5 |
76.5 |
70.5 |
-8.8 |
|
6 |
(Ve) |
p-CH3 |
77.5 |
82.2 |
74.5 |
-9.3 |
|
7 |
(Vf) |
p-F |
54.0 |
32.0 |
58.0 |
-9.1 |
|
8 |
(Vg) |
p-OH |
66.5 |
69.5 |
69.5 |
-9.2 |
|
9 |
(Vh) |
m-Cl |
72.5 |
82.5 |
75.5 |
-8.8 |
|
10 |
(Vi) |
m-Br |
69.0 |
76.0 |
70.0 |
-8.7 |
|
11 |
(Vj) |
m-F |
61.0 |
44.0 |
64.0 |
-9.6 |
|
12 |
(Vk) |
m,p-Cl |
62.5 |
68.4 |
68.5 |
-9.5 |
|
13 |
(Vl) |
m,p-OCH3 |
65.5 |
69.3 |
67.5 |
-9.0 |
|
14 |
(Vm) |
m-NO2 |
80.0 |
83.0 |
82.0 |
-9.1 |
|
15 |
Cisplatin |
– |
23.60 |
24.86 |
29.32 |
– |
LDH discharge examine was performed to test the cell membrane harm incited by Compound Vf. For this, the MDAMB231 cell line was refined with 10, 50, 100, and 250 µM concentration of compound Vf and LDH discharged was estimated at 24, 48, and 72 hrs. Predictable with the above outcomes, a portion and time-subordinate increment in LDH discharge was monitered, and confirming the cytotoxic capability of Compound Vf.(Fig. 1) was confirmed.
 |
Figure 1: LDH assay of compound Vf against MDAMB231 cell line FACS analysis |
FACS investigation was performed to decide if compound Vf has any impact on cell cycle movement. MDAMB231 cells were recolored with propidium iodide after 48 h of treatment 50 and 100 µM and exposed to FACS. Histogram of the vehicle (DMSO) rewarded cells demonstrated a standard cell cycle design, which incorporates G1 and G2 isolated by the S stage. The subG1 step (for the most part dead cells) was not noticeable. Endless supply of compound 1, a subordinate focus change was seen in the cell cycle design (Fig.2,3 and 4)
Compound Vfhas shown S arrest from 19.94% and 32.39%, respectively. G2M phase arrest was found to be 6.38% and 10.44% in MDAMB231 cells. Thus, these outcomes show that compound 1 may meddle with cell division by initiating S stage capture followed by apoptosis. So, more researches are required to comprehend the mechanism of cell cycle capture.
 |
Figure 2: Flow Cytometry Control plots of MDAMB231 cells |
 |
Figure 3: Flow cytometry analysis of cell cycle distribution of MDA-MB-231 cells. The cells were treated with Vf (50μg/mL) for 48 h. Histograms represent sub-G0, G1, S, and G2/M phases of MDA-MB-231 cells. The results were expressed as percentage of total treated cells. Click here to View figure |
 |
Figure 4: Flow cytometry analysis of cell cycle distribution of MDA-MB-231 cells. The cells were treated with Vf (100 μg/mL) for 48 h. Histograms represent sub-G0, G1, S, and G2/M phases of MDA-MB-231 cells. The results were expressed as percentage of total treated cells. Click here to View figure |
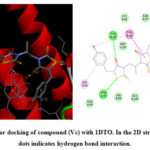 |
Figure 5: Molecular docking of compound (Vc) with 1DTO. In the 2D structure, the green dots indicates hydrogen bond interaction. |
Conclusion
In the present work, fourteen new 5-benzylidene bis-rhodanine derivatives (V) and (Va-m) were have been prepared, characterized by proton NMR, carbon NMR spectroscopy, elemental analysis and evaluated for their anticancer activities against HeLa cell lines by MTT assay, leukemic cell line K562, breast cancer cell line MDAMB231 and molecular docking studies showed better binding scores. Screening of (Vc) and (Vf) compounds was carried for their potential anticancer activity. The results proposed that further study of such compounds with 5-benzylidenebisrhodanine moietymay be interest.
Acknowledgment
The authors acknowledge Central Instruments Facility, Karpagam Academy of Higher Education for providing all the equipments, instruments, chemicals and materials during this research work.
Conflicts of Interest
The authors declares no conflict of interest.
References
- Whiting, E.; Raje, M.R.; Chauhan, J.; Wilder, P.T.; Eker, D.V.; Hughes, S.J.; Bowen, N.G.; Vickers, G.E.A.; Fenimor, I.C.; Fletcher, S.Bioorg. Med. Chem. Lett. 2018,28,523-528.
CrossRef - Mendgen, T.; Steuer, C.; Klein, C.D. J. Med. Chem.2012, 55, 743-747.
CrossRef - Hotta, N.; Akanuma, Y.; Kawamori, R.; Matsuoka, K.; Oka, Y. ; Shichiri, M. ; Toyota, T.; Nakashima, M.; Yoshimura, I.; Sakamoto, N.; Shigeta, Y. Diabetes Care.2006, 29, 1538-1544.
CrossRef - Nagahara, K.; Anderson, J. D.; Kini, G. D.; Dalley, N.K.; Larson, S. B.; Smee, D. F; Jin, A.; Sharma, B. S.; Jolley, W. B. J. Med. Chem.1990, 33, 407-415.
CrossRef - Nazaktabar, A.; Lashkenari, M.S.; Araghi, A.; Ghorbani, M.; Golshahi, H.Int. J. Biol. Macromol.2017, 103, 379-384.
CrossRef - Sortino, M.; Delgado, P.; Juarez, S.; Quiroga, J.; Abonia, R.; Insuasty, B.; Nogueras, M.; Rodero, L.; Garibotto, F. M.; Enriz, R. D.; Zacchino,S. A. Bioorg. Med. Chem.2006, 15, 484-494.
CrossRef - Kartsev, V.; Shikhaliev,K.S.; Geronikaki, A.; Medvedeva, S.M.; Ledenyova, I.V.; Krysin , M.Y.; Petrou, A.; Ciric, A.; Glamoclija, J.; Sokovic, M.Eur. J. Med. Chem.2019, 175,201-214.
CrossRef - Shaikh, M.S.; Kanhed,A.M.; Chandrasekaran, B.; Palkar, M.B.; Agrawal, N.; Lherbet, C.; Hampannavar,G.A.; Karpoormath, R.Bioorg. Med. Chem. Lett. 2019, 29, 2338–2344.
CrossRef - Chao, L.; Jia-Chun, L.; Ya-Ru, L.; Cheng, G.; Mei-Ling, Z.; Hong-Yan, L.; Xiao-Zhen, L.; Chang-Ji, Z.; Hu-Ri, P. Bioorg. Med. Chem. Lett. 2015, 25, 3052-3057.
- Liu,H.; Sun, D.; Du,H.; Zheng, C.; Li,J.; Piao, H.; Li, J.; Sun, L.Eur. J. Med. Chem.2019, 172, 163-173.
CrossRef - Trotsko, N.; Kosikowska, U.; Paneth, A.; Wujec, M.; Malm, A. Saudi. Pharm. J. 2018, 26, 568-577.
CrossRef - Tarahomi , M.; Baharfar, R.; Mohseni, M. Clin. Microbiol. Infect. 2019, 4, 1-5.
CrossRef - Rostam, A.B.; Peyravi, M.; Ghorbani, M. Appl. Surf. Sci.2018,427, 17-28.
CrossRef - Kratky, M.; Vinsova, J.; ikova, J.I.S. Bioorg. Med. Chem. 2017, 25, 1839–1845.
CrossRef - Bhatt, H. B.; Sharma, S.Arab. J. Chem.2013, 10, 1590-1596.
- Abdel Hafez, N.A.; Elsayed, M.A.; El-Shahawi, M.M.; Awad,G.E.A.; Ali,K.A. J. Heterocycl. Chem. 2018, 55, 1729-1737.
CrossRef - Wacothon, K.C.; Ludovic, P.; Anoubilé, B.; Yves-Alain, B.; Rémy, L.G.; Myriam, R.; Anne, C.; Jean, P.B. Med. Chem. Res.2015, 24, 1653-1661.
- Celestina, S.K.; Sundaram, K.; Ravi, S.Bioorg. Chem.2020, 97, 103640.
CrossRef - Khan, N.; Gautam, G.; Gupta, A.k.J. D. D. T. 2019, 9, 161-167.
CrossRef - Andleeb, H.; Tehseen, Y.; Shah, S.J.A,; khan, I.; Iqbal, J.; Hameed, S. RSC Advances.2016, 6(81), 77688–77700.
CrossRef - Sim, M. M.; Ng, S. B.; Buss, A. D.; Crasta, S. C.; Goh, K. L.; Lee, S. K. Bioorg. Med. Chem. Lett.2002, 12, 697-699.
CrossRef - Talele, T. T.; Arora, P.; Kulkarni, S. S.; Patel, M. R.; Singh, S.; Chudayeu, M.; Kaushik-Basu, N. Bioorg. Med. Chem. 2010, 18, 4630-4638.
CrossRef - Ahn, J. H.; Kim, S. J., Park, W. S., Cho, S. Y., Ha, J. D., Kim, S. S., Kang, S. K., Jeong, D. G., Jung, S. K., Lee, S. H., Kim, H. M., Park, S. K., Lee, K. H., Lee, C. W., Ryu, S. E., & Choi, J. K. Bioorg. Med. Chem. Lett. 2006, 16, 2996-2999.
CrossRef - Ahmed I. Khodaira, A.I.; Awadb, M.K.; Gessonc, J.P; Elshaierd, Y.A.M.M.Carbohydr. Res. 2020, 487, 107894.
- Yan, M.; Zhao, J.; Sun, D.; Sun, W.; Wenting, B.Z.; Deng, W.; Zhang, D.; Wang, L. Tetrahedron.Lett.2017, 73(24), 3355–3362.
CrossRef - Dago, C. D.; Ambeu, C.N.T.; Coulibaly, W.K.; Bekro, Y.A.; Bekro, J.A.M.; Guevel, R.L.; Corlu, A.; Pierre Bazureau, J. Chem.Heterocycl. Compd. 2017, 53(3), 341–349.
CrossRef - Moorthy B. T.; Ravi S.; Srivastava M.; Chiruvella K. K.; Hemlal H.; Joy O.; Raghavan S. C. Bioorganic & Medicinal Chemistry Letters. 2010, 20, 6297-6301.
CrossRef - Ravi, S.; Chiruvella, K. K.; Rajesh, K.; Prabhu, V.; Raghavan, S. C. Eur. J. Med. Chem.2010, 45, 2748-2752.
CrossRef - Didwagh, S.S.; Piste, P.B. J. Chem. Pharm. Res. 2013, 5(5), 271-274.
- Kamila, S.; Biehl, E.R.Tetrahedron.Lett.2012, 53, 3998-4003.
CrossRef - Braddock, D.C.; Cailleau, T.; Cansell, G.; Hermitage, S.A.; Pouwer, R.H.; Redmond, J.M.; White, A.J.P. Tetrahedron. Asymmetry.2010, 21, 2911-2919.
CrossRef - Arafa,W.A.A.; Shaker, R.M.; Rabeh, S.A. Heterocycles.2016, 92, 1224-1243.
CrossRef - Harshitharaj, P.K.; Kumar, R.; Ravi, S. J. chem. pharm. sci.2016, 9 (4), 2478-2482.

This work is licensed under a Creative Commons Attribution 4.0 International License.









