Formation Equilibria of Ternary Complexes of Co(Ii) with Girard T (Gt) and Some Amino Acids in Aqueous Solution
Sameerah I. Al-Saeedi1,2 , Reda A. Ammar1,2and Abdel-Nasser M. A. Alaghaz2,3
1Department of Chemistry, College of Science, Princess Nourah Bint Abdul Rahman University
2Deanship of Scientific Research, Princess Nourah Bint Abdul Rahman University, Riyadh, Saudi Arabia
3Department of Chemistry, Faculty of Science, Jazan University, Jazan, Saudi Arabia
Corresponding Author E-mail: raammar@pnu.edu.sa
DOI : http://dx.doi.org/10.13005/ojc/320309
Article Received on : April 27, 2016
Article Accepted on : May 30, 2016
Equilibrium studies have been carried out on complex formation of Co(II), with Girard T (GT) and L = amino acids in aqueous solution, at 25◦C and ionic strength of I = 0.1M (NaNO3). The ternary complexes of Co(II) are formed by stepwise mechanism. The concentration distribution of the complexes is evaluated.
KEYWORDS:pH-titration; Ternary complexes; Girard T; Amino acids
Download this article as:| Copy the following to cite this article: Al-Saeedi S. I, Ammar R. A, Alaghaz A. M. A. Formation Equilibria of Ternary Complexes of Co(Ii) with Girard T (Gt) and Some Amino Acids in Aqueous Solution. Orient J Chem 2016;32(3). |
| Copy the following to cite this URL: Al-Saeedi S. I, Ammar R. A, Alaghaz A. M. A. Formation Equilibria of Ternary Complexes of Co(Ii) with Girard T (Gt) and Some Amino Acids in Aqueous Solution. Orient J Chem 2016;32(3). Available from: http://www.orientjchem.org/?p=17284 |
Introduction
Recently there has been considerable interest in the study of binary, ternary and quaternary complexes by pH-metric method (1-4). Ternary metal complexes in presence of organic and inorganic ligands, amino acids, peptides, proteins, vitamins and enzymes have been recently studied due to their ability as metal systems for metal–protein complexes such as metallo-enzymes. They received particular attention and have been employed in mapping protein surfaces(5), as probes for biological redox centers(6) and in protein capture for both purification(7) and study (8,9). Study of the structure of model ternary complexes provides information about how biological systems achieve their specificity and stability, as well as strategies to improve these features for biotechnological applications. Girard’s reagents have been for a long time used in organic chemistry for the purpose of separating carbonyl compounds from various complex mixtures in the form of water soluble hydrazones (10). Among them, Girard-T (trimethylacetylhydrazide ammonium chloride), [HGT]Cl, (Fig. 1). The interest in the complexes of these reagents and their hydrazones, which, like the other Schiff bases, contain two or more ligating atoms, is of a more recent date. In the known complexes involving hydrazones of Girard’s reagents, the bidentate (11-13), or more frequently, tridentate (14–17) character has been assigned mostly on the basis of spectral data. Girard-T complexes of divalent metal ions M(II) have been reported in solid state (18), but it’s in aqueous solutions are very few . Such complexes are very interesting from both the chemical and biological point of view (19). In the present investigation we describe the equilibria associated with the formation of binary and ternary complexes involving Co(II), Girard T and some amino acids by using potentiometric method at (I = 0.1M NaNO3 -25°C).
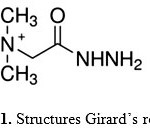 |
Figure 1: Structures Girard’s reagent T |
Experimental
Materials and Reagent
Girard-T (GT) was obtained from Aldrich. Their solutions were prepared and their concentrations were determined by conventional chemical methods (20) . The amino acids and related compounds, Glycine, В-phenylalanine, Valine, Serine, Methylamine, Methionine, S-methylcysteine, Histidine, Histamine, Lysine, Ornithine, Cysteine, Pencillamine and Mercaptoethylamine were provided by Sigma. All these chemical were used as received without any further purification, their purities ranged from 99–99.9%. Co(NO3)2.6H2O was provided by BDH-Biochemicals Ltd. Carbonate-free NaOH (titrant) was prepared and standardized against potassium hydrogen phthalate solution. All solutions were prepared in deionized water.
Apparatus and Measuring Techniques
The pH-measurements were performed with a Metrohm 686 titroprocessor equipped with a 665 Dosimat (Switzerland-Herisau). The electrode was calibrated with standard buffer solutions prepared according to NBS specifications (21). The pH meter readings were converted in to hydrogen ion concentration by titrating a standard acid solution (0.05 M), the ionic strength of which was adjusted to 0.1 M (NaNO3) with a standard base solution (0.05 M) at 25°C. The pkw of water was calculated at ionic strength of 0.1 M to be 13.87±0.05.
Equilibrium Measurements
The following mixtures [A-D] were prepared and titrated potentiometrically with (0.05M) NaOH solution to investigate the equilibria involved in proton-and/or metal ion-ligand complexes.
A. 40 ml of a solution containing (1.25×10−3 M) of GT (GT= girard-T) as primary ligand and 0.1M NaNO3.
B. 40 ml of a solution containing (1.25×10−3 M) of L (L = amino acid) as secondary ligand and 0.1M NaNO3.
C. 40 ml of a solution containing (1.25×10−3 M) Co(II) solution, (2.5× 10−3 M) GT or L and 0.1M NaNO3.
D. 40 ml of a solution containing (1.25×10−3 M) Co (II) solution, (1.25×10−3 M) GT, (1.25×10−3 M) L and 0.1M NaNO3.
The protonation constants of GT (or L) were determined by titration of mixture (A) and (B). The formation constants of Co(II)- GT and Co(II)-L complexes were determined by titration of mixture (C). The respective stability constants of Co(II)- GT -L for the ternary complexes were determined by titration of mixture (D). For all the titrations, HNO3 solution was added, so that they were fully protonated at the beginning of the titrations.
Computer Analysis of Titration Data
Calculations were performed using the computer program SUPERQUAD(22). The stoichiometrics and stability constants of the complexes formed were determined by trying various possible composition models for the systems studied. The model selected was that which gave the best statistical fit and which was chemically consistent with the titration data without giving any systematic drifts in the magnitudes of various residuals, as described elsewhere (23). The species distribution diagrams were obtained with the program SPECIES (24).
Results and Discussion
The acid dissociation constants of the amino acids ligands and the formation constants of their binary complexes were previously reported (25). We have redetermined these constants, Table (1) under the prevailing experimental conditions as those utilized for determining the stability constants of the mixed-ligand complexes.
Acid-Base Equilibria Of Gt.
The calculated dissociation constant of girard T (GT) are given in (Table 1). The highest pKa value due to the dissociation of the proton in the methylene group located between a carbonyl and a quaternary ammonium group in view of the electron attracting effect of the quaternary ammonium group
Binary Co (II) Complex Formation Equilibria with GT.
Different equilibrium models have been tried to acquire best fit with the experimental potentiometric data for Co(II)-GT system. The selected model with the best statistical fit was found to consist of 110, 120 and 11-1 species. The stability constants of their complexes are given in Table (1). Previous studies(26) on related systems favoured the occurrence of structure (I) for 110 complex and reported the following scheme for acid dissociation from 110 complex yielding structure (II) which is referred to as 11-1.
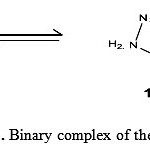 |
Scheme 1: Binary complex of the Girard-T |
The concentration distribution diagram of Co-GT system is shown in Fig.(2). 110 complex starts to form at pH = 1 and its concentration reaches a maximum of about 99.84 at pH = 5.2. The concentration of 120 complex increases with increasing pH and reaches a maximum of about 43.37% at pH 9.2. The deprotonated complex, 11-1 starts to form at pH=6.6 and reaches a maximum of about 98.12% at pH= 11.8.
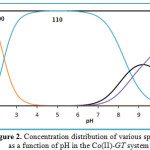 |
Figure 2: Concentration distribution of various species as a function of pH in the Co(II)-GT system |
Ternary Co(II) Complex Formation Equilibria Involving Girard T (GT) and Amino Acids.
The potentiometric titration curves associated with the formation of Co(GT) complex in absence and presence of a secondary ligand (amino acids) are shown in Fig. (3), Co(II)-GT–S-methylcysteine complex, taken as a representative. The formation of a ternary complex has been ascertained by comparing the mixed ligand titration curve with the composite curve obtained by graphical addition of the secondary ligand titration data to the Co(GT) titration curve. The mixed-ligand titration curve was found to deviate considerably from the calculated composite curve, indicating the formation of a ternary complex. It may be plausibly assumed that in the presence of both ligands, GT would first be ligated to the Co(II) ion, followed by ligation of the secondary ligand (L), whereby the formation of the ternary complex may be conceived as a stepwise process involving the equilibria:
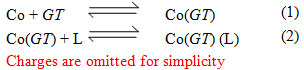
The results (Table 1) show that the amino acids form 1110 complexes but methylamine forms 1110 and 1120 complexes. The methylamine complex (1110) is less stable than those of amino acids. This indicates that amino acids function as bidentate ligands coordinating through the amino and carboxylate groups. The stability constant values of methionine and S-methylcysteine complexes are in fair agreement with those of a-amino acids and lower than that of mercaptoethylamine (N,S-donor set). This may be taken to indicate that methionine and S-methylcysteine chelate through the a-amino and carboxylate groups.
But for sulphur ligands, penicillamine, cysteine and mercaptoethylamine, the ternary complex formation proceed through simultaneous mechanisms depending on the chelating potential of GT and other ligands. The formation constants of the 1:1 Co(II) complexes with GT and those of sulphur ligands (Table1), are of the same order. Consequently, the ligation of GT and sulphur ligands will proceed simultaneously.
The complexes formed can be characterized with the following general equilibrium process and stability constant:
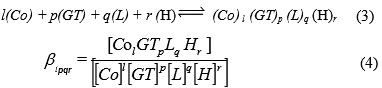
The stability constants defined by Eq. (4) were calculated from the pH-metric titration curves in the usual way with the aid of the computer program SUPERQUAD(14).
Computer analysis of the pH titration data revealed the formation of the ternary complexes Co-GT-L and the corresponding protonated species Co(II)-GT-LH. The acid dissociation constants of the protonated ternary complex (logβ1111–logβ1110) obtained with penicillamine, and mercaptoethylamine are 6.46, and 5.58 respectively. These values obtained in the present study are less than the previously reported microscopic acid dissociation constants (27) of [–NH3]+ and –SH groups of penicillamine, cysteine and mercaptoethylamine, revealing that the [–NH3]+ and –SH groups most likely take part in complex formation.
The relative stability of the ternary complexes formed through stepwise mechanism, as compared to those of the corresponding binary complexes, is expressed in terms of ∆ log K as defined by the following equation:
![]()
The ∆ log K values given in Table 1 are invariably negative, showing that the amino acids (L–) form more stable complexes with Co(II)-GT than with Co(II)
Estimation of the concentration distribution of the various species in solution is a useful mean for elucidating the extent of Co(II) binding capacity toward the primary and secondary ligands. The concentration distribution of the ternary complex formed with amino acids attained values ranging from about 88% to 99.9% along the pH range 7.8-10.6. Protonated ternary complex species have been found to be most favoured at lower pH values. The species distribution for Co-GT– Ornithine complex, taken as a representative, is given in Fig. (4).
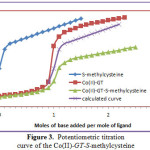 |
Figure 3: Potentiometric titration curve of the Co(II)-GT–S-methylcysteine |
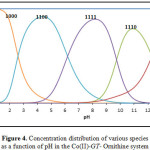 |
Figure 4: Concentration distribution of various species as a function of pH in the Co(II)-GT– Ornithine system |
Table 1: The Acid Dissociation Constants Of The Ligands And The Formation Constants Of The Binary And Ternary Complexes In The Co(II)- GT Or Amino Acids Systems At 25◦C And 0.1 M Ionic Strength.
|
System
|
l |
p |
q |
ra |
logВ b |
∆ log K |
||||||
|
Girard-T (GT) |
0 0 1 1 1 |
1 1 1 2 1 |
0 0 0 0 0 |
1 2 0 0 -1 |
9.71(0.006) 12.51(0.01) 10.62(0.01) 14.92(0.05) 1.94(0.05)
|
|||||||
|
Glycine |
0 0 1 1 1 |
0 0 0 0 1 |
1 1 1 2 1 |
1 2 0 0 0 |
9.62(0.02) 11.90(0.01) 5.78(0.04) 10.23(0.03) 5.56(0.01)
|
-0.22 |
||||||
|
В-phenylalanine |
0 0 1 1 1 |
0 0 0 0 1 |
1 1 1 2 1 |
1 2 0 0 0 |
9.12(0.01) 11.01(0.03) 4.75(0.01) 8.66(0.02) 4.11(0.05) |
-0.64 |
||||||
|
Valine |
0 0 1 1 1 |
0 0 0 0 1 |
1 1 1 2 1 |
1 2 0 0 0 |
9.57(0.02) 11.70(0.01) 5.41(0.03) 9.72(0.04) 5.09(0.03) |
-0.32 |
||||||
|
Serine
|
0 0 1 1 1 1 |
0 0 0 0 1 1 |
1 1 1 2 1 1 |
1 2 0 0 0 -1 |
9.14(0.05) 11.40(0.01) 5.51(0.03) 10.11(0.04) 5.20(0.04) -3.71(0.07)
|
-0.31 |
||||||
|
Methylamine
|
0 1 1 1 1 |
0 0 0 1 1 |
1 1 2 1 2 |
1 0 0 0 0 |
10.55(0.01) 4.44(0.01) 9.87(0.03) 4.00(0.06) 8.12(0.04) |
-0.44 |
||||||
|
System
|
l |
p |
q |
ra |
logВ b |
∆ log K |
||||
|
Methionine |
0 0 1 1 1 |
0 0 0 0 1 |
1 1 1 2 1 |
1 2 0 0 0 |
9.12(0.01) 11.10(0.04) 5.23(0.02) 9.78(0.04) 5.10(0.02) |
-0.23 |
||||
|
S-methylcysteine |
0 1 1 1 |
0 0 0 1 |
1 1 2 1 |
1 0 0 0 |
8.25(0.01) 5.39(0.02) 10.11(0.03) 5.21(0.03) |
-0.18 |
||||
|
Histidine |
0 0 0 1 1 1 1
|
0 0 0 0 0 0 1
|
1 1 1 1 2 1 1
|
1 2 3 0 0 1 0
|
9.53(0.01) 15.81(0.03) 17.81(0.06) 7.87(0.04) 14.01(0.06) 19.76(0.03) 6.98(0.05)
|
-0.89 |
||||
|
Histamine |
0 0 1 1 1 1 |
0 0 0 0 1 1 |
1 1 1 2 1 1 |
1 2 0 0 0 1 |
9.85(0.01) 16.05(0.05) 6.81(0.02) 11.33(0.03) 6.13(0.02) 13.33(0.05) |
-0. 68 |
||||
|
Lysine |
0 0 1 1 1 1 |
0 0 0 0 1 1 |
1 1 1 2 1 1 |
1 2 0 0 0 1 |
10.44(0.01) 19.66(0.01) 7.56(0.02) 11.24(0.03) 6.98(0.03) 15.12(0.04) |
-0.58 |
||||
|
Ornithine |
0 0 0 1 1 1 1 1 |
0 0 0 0 0 0 1 1 |
1 1 1 1 2 1 1 1 |
1 2 3 0 0 1 0 1 |
10.58(0.00) 19.43(0.02) 21.39(0.02) 7.31(0.02) 11.02(0.02) 15.13(0.04) 6.15(0.04) 14.73(0.03) |
-1.16 |
||||
|
System
|
l |
p |
q |
ra |
logВ b |
∆ log K |
||
|
Cysteine |
0 0 1 1 1 1 |
0 0 0 0 1 1 |
1 1 1 2 1 1 |
1 2 0 0 0 1 |
9.80(0.01) 17.70(0.05) 9.75(0.03) 19.55(0.06) 19.83(0.03) 25.97(0.05) |
-0.54 |
||
|
Pencillamine |
0 0 1 1 1 1 |
0 0 0 0 1 1 |
1 1 1 2 1 1 |
1 2 0 0 0 1 |
10.10(0.01) 17.97(0.02) 10.68(0.05) 2.31(0.009) 20.23(0.01) 26.89(0.05) |
-1.07 |
||
|
Mercaptoethylamine |
0 0 1 1 1 1 |
0 0 0 0 1 1 |
1 1 1 2 1 1 |
1 2 0 0 0 1 |
10.03(0.04) 18.64(0.02) 10.78(0.04) 18.23(0.02) 21.02(0.01) 26.00(0.03) |
-0.38 |
||
a l, p, q and r are the stoichiometric coefficient corresponding to Co(II), GT, amino acids and H+ respectively, bstandard deviations are given in parentheses.
Conclusion
The present investigation describes the formation equilibria of Co(II) complexes involving Girard-T (GT) as primary ligand and some amino acids as secondary ligands in aqueous media by using potentiometric titration method at (I = 0.1M NaNO3 -25°C). The complexation behavior of Co)II) with (GT) and amino acids shows the formation of ternary complexes in a stepwise manner in aqueous media. The relative stabilities of the ternary complexes are compared with those of the corresponding binary complexes in terms of Δ log K values . Additionally, species distributions in solution for all complexes were precisely evaluated. pH-studies of these systems clearly indicate that the concentration of the ternary complexes increases with increasing pH. This behaviour attests that complex formation is more favoured in physiological pH range (6-7). The ternary complexes formed may have interesting biological activity and more research work need to be conducted by biologists.
Acknowledgment
This research project was supported by a grant from the Deanship of Scientific Research. Princess Nourah Bint Abdul Rahman University.
Reference
- R. A. Ammar, N. A. Alarfaj and M. F. El-Tohamy, Int. J. Electrochem. Sci., 7 (2012) 1512 .
- W.M. Hosny, S.M. El-Medani and M.M. Shoukry, Talanta, 48 (1999) 913.
CrossRef - S. Singh, Transition Metal Chemistry, 25(2000) 9.
CrossRef - A. A. El-Sherif, J. Coord. Chem, 64 ( 2011) 1240.
CrossRef - J.R. Bocarsly and J.K. Barton, Inorg. Chem., 31(1992) 2827.
CrossRef - O. Farver and I. Pecht, Coord. Chem. Rev., 95 (1989) 17.
CrossRef - J. Crowe, H. Dobeli, R. Gentz, E. Hochuli, D. Stuber and K. Henco, Methods Mol. Bio., 31(1994) 371.
- S. E. Nieba-Axmann, A. Persson, M. Hamalainen, F. Edebratt, A. Hansson, J. Lidholm , K. Magnusson, A.F. Karlsson and A. Pluckthun, Anal. Biochem., 252 (1997) 217.
CrossRef - K.M. Maloriery, D.R. Shnek, D.Y. Sasaki and F.H. Arnold, Chem. Biol., 3 (1996) 185.
CrossRef - A. Girard and G. Sandulesco, Helv. Chim. Acta, 19 (1936) 1095.
CrossRef - M.E.M. Emam, M.A.H. Hafez and M.N.H. Moussa, J. Therm. Anal., 37 (1991) 1005.
CrossRef - M.M. Mostafa, M.A. Khattab and K.M. Ibrahim, Transition Met. Chem., 8 (1983) 212.
CrossRef - M.M. Mostafa, S.M. Hassan and G.M. Ibrahim, J. Inorg. Nucl. Chem., 42 (1980) 285.
CrossRef - S.M. Al-Ashqar and M.M. Mostafa, Spectrochim. Acta, Part A 71 (2008) 1321.
CrossRef - M.M. Abou Sekkina and M.R. Salem, J. Therm. Anal. 48 (1997) 841.
CrossRef - R.M. El-Bahnasawy, J. Therm. Anal. 45 (1995) 1547.
CrossRef - V.M. Leovac, K. Mészáros-Szécsényi, L. j. S. Vojinović-Ješić, V. I. Češjević, S. Markov and T. Wadsten, J. Therm. Anal. Calorim., 86 (2006) 379.
CrossRef - M. Masui and H. Ohmori, J. Chem. Soc., A (1969) 153
CrossRef - A. M. A. El-Sokkary, M. M. El-Naggar and A. F. Abdel-Aziz, Appl. Organomet. Chem., 24 (2010) 439
- Vogel, 1989. Text Book of Quantitative Chemical Analysis, 5th edn. Longman, Harlow, 326, Chap. 10.
- R.G. Bates, 1975. Determination of pH—Theory and Practice, 2nd edn.Wiley-Interscience, New York.
- P. Gans, A. Sabatini and A. Vacca, J. Chem. Sot. Dalton Trans., 22(1985)1195.
CrossRef - P. Gans, A. Sabatini and A. Vacca, Inorg. Chim. Acta, 18, (1976)237.
CrossRef - L. Pettit, University of Leeds. Personal communication (1984).
- D.D. Perrin, Stability Constants of Metal-Ion Complexes, Part B; Organic Ligands. Pergamon, Oxford, ( 1979).
- M.M. Mostafa, S.M. Hassan and G.M. Ibrahim, J .Iong. Nucl. Chem., 42 (1980) 285.
CrossRef - R. S. Reid and D. L. Rabenstein, Can. J. Chem., 59 (1981)1505.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.









