Recombinant Triplex formed by PNA-TFO: A Molecular Dynamics Simulation Study
Vijaya Shri Mall , Rajendra Prasad Ojha and Rakesh Kumar Tiwari
, Rajendra Prasad Ojha and Rakesh Kumar Tiwari
D. D. U. Gorkahpur University, Gorakhpur India.
Corresponding Author E-mail: vijayashrimall@gmail.com
DOI : http://dx.doi.org/10.13005/ojc/340528
Article Received on : 22-06-2018
Article Accepted on : 14-08-2018
Article Published : 28 Sep 2018
Building of high affinity triplex-forming oligonucleotides(TFOs) enhances its therapeutical application. Peptide nucleic acid(PNA), a modified DNA oligomer with neutral backbone enhances the affinity of TFO. MD simulation method is very helpful to study the stability, affinity and behavior of complex at nanosecond scale. Therefore a 15-mer PNA-TFO is used here to model DNA:DNA:PNA triplex in mixed purine/pyrimidine sequence. DNA:DNA:DNA triplex, DNA:DNA duplex and DNA:PNA duplex were also modeled for comparison. 100ns of MD run on all four complexes in solution at neutral pH. The triplex conformation stabilized with Recombinant type(R-type) of Hydrogen bonding during simulation. The rmsd of DNA:DNA:PNA triplex and DNA:DNA:DNA triplex converges after 45 ns of dynamics and the binding affinity of PNA-TFO found greater than DNA-TFO. Together with non-toxicity of PNA oligomer, stable triplex formation with R-type of hydrogen bonding pattern and high binding affinity in mixed sequence promotes the study regarding Recombinant triplex with PNA-TFO.
KEYWORDS:DNA; PNA; R-type; TFO; Triplex
Download this article as:| Copy the following to cite this article: Mall V. S, Ojha R. P, Tiwari R. K. Recombinant Triplex formed by PNA-TFO: A Molecular Dynamics Simulation Study. Orient J Chem 2018;34(5). |
| Copy the following to cite this URL: Mall V. S, Ojha R. P, Tiwari R. K. Recombinant Triplex formed by PNA-TFO: A Molecular Dynamics Simulation Study. Orient J Chem 2018;34(5). Available from: http://www.orientjchem.org/?p=49932 |
Introduction
Triplex-forming Oligonucleotides (TFOs) are short oligonucleotides binds in major groove of double helical DNA and form DNA triplex. The triplex formation play important role in most of the vital function as in replication, transcription, homologous recombination, gene regulation etc. A lot of therapeutical application discussed in several reviews.1,2 The formation of triple helical structure of DNA is straight forward in purine/pyrimidine rich sequence in-vitro. But the nuclear environment produces obstacle in formation of triplex in-vivo, in terms of mixed sequence of DNA, low binding affinity of TFO to the duplex DNA, due to unfavorable electrostatic repulsion between the three negatively charged strands.3, 4 The studies regarding triplexes commonly based on Hoogsteen/reverse Hoogsteen triplexes, where the TFO recognizes one of the strands of DNA duplex. But the studies related to the Recombinant triplex(R-triplex), where TFO recognizes both the bases coming from double helical DNA are very few.5 The formations of Hoogsteen/reverse Hoogsteen triplexes are limited to purine/pyrimidine rich sequences.6 The R-triplex formation is much suitable to natural environment of nucleic acids, can efficiently form triplex in even in mixed sequence of duplex DNA.7 To overcome these limitations of triplex formation modification in TFO came into account. Peptide Nucleic Acids (PNA) is artificially designed oligonucleotide with neutral backbone, a mimic of DNA shows Watson-Crick pattern when synthesize with DNA, RNA or PNA itself.8 PNA oligomer also show sequence specificity when hybridized as Hoogsteen/reverse Hoogsteen triplex with purine/pyrimidine rich sequences.9 PNA-TFO is thus may efficiently form R-triplex in mixed sequence when hybridized with mixed sequence DNA duplex.10,11 Molecular Dynamics simulation methods are very useful in the study of complex behavior, affinity, stability and Hydrogen bonding pattern in the solution and other environment in-vitro. Present work is comparative MD study regarding stability, affinity and structural conformation of R-triplex formed by PNA-TFO and DNA-TFO in mixed sequence of DNA duplex. The sequence 5’-AGGCCGGACCCGGCG-3’ is mixed purine/pyrimidine 15-mer sequence is used to design all the complexes. Triplex formed by PNA-TFO and DNA-TFO is denoted as DNA:DNA:PNA triplex and DNA:DNA:DNA triplex respectively. In addition, two more studies has been done with DNA:DNA duplex and DNA:PNA duplex in the same 15-mer sequence.
Computational Methods
Triplex modeling and simulation setup
The initial structure of 15-mer DNA:DNA:PNA triplex of sequence 5’-d(AGGCCGGACCCGGCG)d(CGCCGGGTCCGGCCT)-PNA(AGGCCGGACCCGGG)-3’ were modeled by using Nucleic Acid Builder of AMBER 1412 and CHIMERA.13 DNA:DNA:DNA triplex, DNA-DNA duplex and DNA-PNA duplex with same sequence were also modeled. Na+ ions were used to neutralize the system and TIP3P.20 BOX water molecule is used to solvate the complex in buffer of 10.0 Angstrom. The box dimension, volume, mass, number of atom, residue and molecule for each complex are given in Table 1.
Table 1: Table for the box dimension, mass, number of atom, residue, molecule and density of each designed complex by LEaP.
| ComplexBox dimension | DNA:DNA duplex | DNA:PNA duplex | DNA:DNA:DNA triplex | DNA:DNA:PNA triplex |
| X (Angstrom) | 45.963 | 45.556 | 53.760 | 45.963 |
| Y (Angstrom) | 47.144 | 47.144 | 54.806 | 47.144 |
| Z (Angstrom) | 80.400 | 80.400 | 79.589 | 80.440 |
| Volume (A3 ) | 174216.7 | 172675.4 | 234501.4 | 234071.5 |
| Mass (amu) | 79582.5 | 72164.9 | 110151.5 | 108924.9 |
| Number of atom | 12622 | 12684 | 17409 | 17315 |
| Residue | 3941 | 3946 | 5403 | 5356 |
| Molecule | 3913 | 3916 | 5361 | 5312 |
| Density (g/cc) | 0.759 | 0.694 | 0.780 | 0.773 |
The coordinates of each complex were taken from Protein data bank. The force-field parameters as angles, dihedrals, improper rotation and charges for PNA were generated and optimized by Gaussian-0315 program using basis-set 6-31G**16
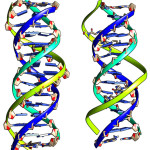 |
Figure 1: DNA:DNA:DNA triplex(right) and DNA:DNA:PNA triplex’ Blue and Cyan color strands are Watson Crick Duplex and TFOs are in with green color strands. Click here to View figure |
Minimization and Equilibration
The structure were minimized by using steepest decent method17 for 500 steps and generalized born method18 for next 500 steps with strong restraint dynamics to weak restraint dynamics in three stages. A weak restraint is used in heating and equilibration to avoid end base fraying apart. The heating is done in six steps and equilibrated in five steps. The equilibration of complexes starts with strong restraint dynamics to zero restraint dynamics. Now the complex is ready for molecular dynamics.
Molecular Dynamics of complexes in solution
The molecular dynamics of all four complexes in TIP3P water solvent starts with the coordinates obtained after equilibration. These dynamics performed under constant pressure periodic boundary condition at constant atmospheric pressure.19 Bonds involving hydrogen were neglected. The molecular dynamics coordinate file will be written after each 2500 steps at time-interval 0.002 ps at constant temperature of 300.0 K. In the same manner 100 ns dynamics performed with AMBER 14 code. The Molecular dynamics movie is recorded to see the variation during dynamics for each simulation. The structure of complex during simulation is shown in figure 1.
Binding Free Energy Calculation
The binding free energy of PNA-TFO to the DNA:DNA duplex, DNA-TFO to the DNA:DNA duplex, DNA oligomer to its complementary DNA and DNA oligomer to its complementary mimic PNA was calculated by MMPBSA/GBSA method21 of AMBER 14 code. The total binding free energy of complex is equal to the sum of gas phase contribution and solvation energy contribution. The relation is given by following equation:
∆Gtotal = ∆Egas + ∆Esolvation −T∆S
Here ∆Egas is given by:
∆Egas = ∆Einternal + ∆Evan_der_Waals + ∆Eelectric
Where ∆Einternal is the internal energy corresponds to bond energy, angle energy and torsional energy terms in the molecular mechanics force field; ∆Evan_der_Waals is the van der Waals energy term; ∆Esolvation is solvation energy term corresponds to the role of complex is solution; ∆Eelectric is the electric energy term corresponds to the contribution of charge.
And binding energy of ligand (TFO) to its receptor (DNA duplex) is the difference of free energy of ligand and receptor to its complex counterpart.
Results
Molecular Dynamics and hydrogen bonding pattern
From the Figure 1, it is seen that the triplex with PNA-TFO covers minimum area than those of triplex with natural DNA-TFO. During the solution dynamics of DNA: DNA: DNA triplex, DNA: DNA: PNA triplex, DNA:DNA duplex and DNA: PNA duplex, it is seen that the electrically neutral peptide backbone are more flexible than those of negatively charged Phosphate backbone in their complexes. Therefore it is expected that PNA backbone should be able to fit the structure with more affinity than those of natural DNA or RNA backbone.20 PNA oligomer follow Watson-Crick pairing rule in formation of duplex and show sequence specificity when used as TFO to recognized duplex DNA.22 Recombinant type of Hydrogen bonding formed between the bases of TFOs and duplex DNA.23 The R-type of hydrogen bonding pattern for A*T-A Triplet, G*C-G Triplet and C*G-C Triplet are maintained during dynamics (figure 2).24, 24 Figure 2 show the third base coming from TFO recognizes both the bases of DNA duplex in major groove.25
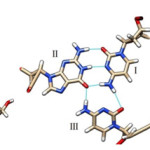 |
Figure 2: (left) Hydrogen Bonding Pattern of G*C-G Triplet with R-Type of Bonding Pattern; the Third Base Guanine Binds G-C Base Pair by Forming R-Type of H-Bonds. |
(Middle) Hydrogen Bonding Pattern of G*C-G Triplet with R-Type of Bonding Pattern; the Third Base Cytosine Binds G-C Base Pair by Forming R-Type of H-Bonds.; (Left) Hydrogen Bonding Pattern of A*T-A Triplet with R-Type of Bonding Pattern; the Third Base Adenine Binds A-T Base Pair by Forming R-Type of H-Bonds.
The root-mean-square-deviation (rmsd)
The rmsd over the course of simulation is used to measure the conformational stability of the complex during simulation. The rmsd plots of DNA:DNA:DNA triplex and DNA:DNA:PNA triplex is shown in figure 3 and comparative rmsd of DNA:DNA duplex and DNA:PNA duplex are plotted in figure 4. The rmsd of DNA:DNA duplex and DNA:PNA duplex are almost same and stabilize between 2.5 to 3.5 Angstrom. The convergence of rmsd observed after 45ns for both the triplex, the rmsd of DNA:DNA:DNA triplex converges near 2.25 Angstrom and DNA:DNA:PNA triplexes converges near 6 Angstrom. Flexible nature of PNA-TFO helps it to get a suitable position in major groove of DNA and RNA backbone14 As a result the triplex conformations stabilize into the form of R-triplex after 45ns of dynamics.
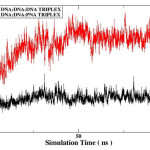 |
Figure 3: Root mean square deviation of DNA:DNA:DNA triplex (black) and DNA:DNA:PNA triplex(red). |
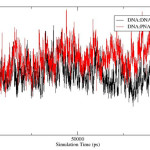 |
Figure 4: Root-mean-square-deviation of DNA:DNA duplex (black) and DNA:PNA duplex(red). |
Binding Free Energy
The binding free energies are used to study the binding of TFO to the DNA duplex and to provide quantitative estimate of their stability.26 To calculate the binding free energy of DNA-DNA duplex; DNA:DNA duplex is treated as complex, the second strand is taken as ligand, the first strand is treated as receptor. For DNA:PNA duplex; DNA:PNA duplex is treated as complex, the second strand (PNA oligomer) is taken as ligand, the first strand (DNA oligomer)is treated as receptor. Thus binding affinity of DNA and PNA to the natural DNA oligonucleotide with Watson-Crick pairing has been calculated. For DNA:DNA:DNA/DNA:DNA:PNA triplexes; the triplex is treated as complex, the DNA:DNA duplex is taken as receptor and TFOs are taken as ligand. Thus the binding affinity of TFO with peptide and natural backbone to the DNA duplex with Recombinant-bonding has been calculated. The corresponding total free energy together and binding free energy for each complex is shown in (Table 2) and in (Table 3). Here the binding free energy of PNA-TFO to the DNA duplex is -34.8733 Kcal/mol and that of DNA-TFO is +94.5075 Kcal/mol. These result shows that natural DNA-TFO does not binds with the DNA duplex to form stable DNA triplex while when it is modified backbone as PNA it forms a stable triplex. The same calculation was done by GBSA methodology, the result is shown in (Table 4) and (Table 5). The result support the result came from GBSA methods as the binding energy of PNA oligomer is high as compared to DNA oligomer together in DNA duplex and DNA triplex.
Table 2: Different Component of Binding Free Energy of Complexes (Complex-receptor-ligand) calculated by Poisson Boltzmann methodology (in Kcal/mol).
| Energy component | DNA-DNA duplex | DNA-PNA duplex | DNA: DNA: DNA triplex | DNA: DNA: PNA triplex |
| Van der waals | -77.7852 | -95.7625 | -103.1164 | -112.3859 |
| Electrical | 2843.4671 | -420.3861 | 6198.7392 | -263.4766 |
| Poisson Boltzmann | -2907.8994 | 292.6126 | -6100.8971 | 262.5199 |
| ENPOLAR | -56.3562 | -68.6443 | -59.6940 | -71.3635 |
| EDISPER | 106.3982 | 132.0922 | 141.5060 | 149.8327 |
| Delta G gas | 2765.6819 | -516.1487 | 6095.6228 | -375.8625 |
| Delta G solvation | -2857.8574 | 356.0606 | -6019.0851 | 340.9892 |
| Delta G total | -92.1755 | -160.0881 | +76.5376 | -34.8733 |
Table 3: Table for the free energy and binding free energy (in Kcal/mol): PBSA Methodology.
|
Molecule |
Complex |
Receptor |
Ligand |
Delta G TOTAL |
|
DNA:DNA duplex |
-3563.9611 |
-1734.2550 |
-1737.5306 |
-92.1755 |
|
DNA:PNA duplex |
-2947.5400 |
-1765.7173 |
-1021.7346 |
-160.0881 |
|
DNA:DNA:DNA triplex |
-5161.4892 |
-1715.7236 |
-3540.2731 |
+94.5075 |
|
DNA:DNA:PNA triplex |
-4316.8827 |
-3620.0519 |
-661.9575 |
-34.8733 |
Table 4: Different Component of Binding Free Energy(Complex-receptor-ligand) calculated by Generalized Born methodology(in Kcal/mol).
| Energy component | DNA-DNA duplex | DNA-PNA duplex | DNA:DNA:DNA triplex | DNA:DNA:PNA triplex |
| Van der waals | -77.7852 | -95.7625 | -103.1164 | -112.3859 |
| Electrical | 2843.4671 | -420.3861 | 6198.7392 | -263.4766 |
| Generalized Born | -2901.0338 | 342.9120 | -6141.1405 | 286.0135 |
| Surface | -8.5599 | -10.1562 | -11.7345 | -12.4878 |
| Delta G gas | 2765.6819 | -516.1487 | 6095.6228 | -375.8625 |
| Delta G solvation | -2909.5936 | 332.7558 | -6152.8750 | 273.5257 |
| Delta G total | -143.9117 | -183.3929 | -57.2522 | -102.3368 |
Table 5: Table for the free energy and binding free energy (in Kcal/mol): GBSA Methodology
| Molecule | Complex | Receptor | Ligand | Delta G TOTAL |
| DNA:DNA duplex | -3197.6066 | -1524.1598 | -1529.5351 | -143.9117 |
| DNA:PNA duplex | -2881.6277 | -1575.8938 | -1122.3410 | -183.3929 |
| DNA:DNA:DNA triplex | -4788.1204 | -3195.6573 | -1535.2108 | -57.2522 |
| DNA:DNA:PNA triplex | -4144.0817 | -3281.4877 | -760.2573 | -102.3368 |
Discussion
The binding affinity of natural TFO to the double helical nucleic acid is low and limited to purine/pyrimidine rich sequences when TFO comes to bind with Hoogsteen/reverse Hoogsteen hydrogen bonding. In the Hoogsteen/reverse Hoogsteen Hydrogen bonding, the TFO oriented towards one of the strand of double helical nucleic acids and therefore hydrogen bonds limited to the donor and acceptor coming from bases of one of the strand of double helical DNA. While in Recombinant Hydrogen bonding, the TFO oriented towards the bases of both the strands of double helix and form hydrogen bonds to both the bases of DNA duplex. Thus TFO with R-type of Hydrogen bonding will be isomeric to the bases coming from both the strands of DNA duplex. This is the major reason of stabilization of R-triplexes in mixed sequence and in natural environment. The structural conformation of R-triplexes in mixed sequences was many times experimentally supported by Shchyolkina and coworkers.5, 9 Another major cause of destabilization of triplexes is unfavorable electrostatic repulsion between negatively charged strands and structural conformation of nucleotides. The nucleic acids with electrically neutral backbone in place of negatively charged backbone efficiently reduce such limitation. PNA is one of the better option to overcome such limitation, therefore the study of R-triplex with PNA TFO will be very useful. The results of this study shows that PNA binds with DNA with greater affinity than those of natural DNA and form W/C duplex. PNA-TFO efficiently binds with DNA duplex than those of natural DNA and forms comparatively stable R-triplexes in mixed purine/pyrimidine sequences of DNA. PNA is nontoxic, electrically neutral, does not react with protein,27 and efficiently binds with target nucleic acid.28, 29 PNA-TFO is therefore will be very useful in cancer therapy, regulating gene-expression, site-specific gene editing and other triplex technologies.30
Conclusion
In the Recombinant triplexes, TFO binds in major groove of DNA duplex, in a manner, the third strand bases recognizes both the bases coming from W/C pair. The third strand backbone modified by Peptide backbone, provides electrically neutral oligonucleotides, therefore will be able to overcome the unfavorable repulsion between the three strands originated due to negatively charged strands of triplexes. Thus PNA oligonucleotide may provide more efficient result, when used as TFO in Recombinant manner.
Acknowledgement
Dr. Rakesh Kumar Tiwari acknowledges UGC, New Delhi.
References
- Buske, F. A.; Mattick, J. S. and Balley, T. L. RNA Biology 2011, 8(3), 427-439.
CrossRef - Hu, Y,; Cecconello, A.; Idili, A.; Ricci, F. and Willner, I. Angew. Chem. Inst, Ed. 2017, 56, 15210-15233
- Fox, K. and Brown, T. Quarterly reviews of biophysics. 2005, 38, 311-20. 10.1017/S0033583506004197.
CrossRef - Duca, M.; Vekhoff, P.; Oussedik, K.; Halby, L.; Arimondo, P. Nucleic Acids Research 2008, 36(16),
CrossRef
- Shchyolkina, A. K.; Kaluzhny, D. N.; Borisova, O. F.; Arndt-Jovin, D. J.; Jovin, T. M. and Zhurkin, V. B. J. Biomol. Struct. Dyn. 2016, 34(6), 1298-1306.
CrossRef - Ojha, R. P. Tiwari, R. K. Nucleic Acids Research 2003, 31 6373-6380.
CrossRef - Fox, K. R. and Brown, T. Biochem. Soc. Trans. 2011 39, 629–634
CrossRef - Gupta, A.; Mishra, A.; Puri, N,; J of Biotech. 2017, 259, 148-159
CrossRef - Kaluzhny, D. N.; Timoshin, V. V.; Borisova O. F.; Zhurkin, V. B.; Florentiev, V. L. and Shchyolkina, A. K.; J. Biomol. Struct. Dyn. 2006, 26(3), 301-306.
CrossRef - Egholm, M.; Buchardt, O.; Christensen, L.; Behrens, C.; Freier, S. M.; Driver, D. A.; Berg, R. H.; Kim, S. K.; Norden, B.; Nielsen, P. E. Nature 1993, 365(6446),566-8
CrossRef - Toh, D.K.; Patil, K. M.; Chen, G. J Vis Exp, 2017, 127
- D.A. Case, I.Y. Ben-Shalom, S.R. Brozell, D.S. Cerutti, T.E. Cheatham, III, V.W.D. Cruzeiro, T.A. Darden, R.E. Duke, D. Ghoreishi, M.K. Gilson, H. Gohlke, A.W. Goetz, D. Greene, R Harris, N. Homeyer, S. Izadi, A. Kovalenko, T. Kurtzman, T.S. Lee, S. LeGrand, P. Li, C. Lin, J. Liu, T. Luchko, R. Luo, D.J. Mermelstein, K.M. Merz, Y. Miao, G. Monard, C. Nguyen, H. Nguyen, I. Omelyan, A. Onufriev, F. Pan, R. Qi, D.R. Roe, A. Roitberg, C. Sagui, S. Schott-Verdugo, J. Shen, C.L. Simmerling, J. Smith, R. Salomon-Ferrer, J. Swails, R.C. Walker, J. Wang, H. Wei, R.M. Wolf, X. Wu, L. Xiao, D.M. York and P.A. Kollman AMBER 2014, 2014 University of California, San Francisco.
- Pettersen, E. F.; Goddard, T. D.; Huang, C. C.; Couch, G. S.; Greenblatt, D. M.; Meng, E. C.; Ferrin, T. E.; J Comput Chem. 2004 25(13) 1605-12.
CrossRef - Price, D. J.; Brooks, C. L, J. Chem.Phys.,2004 121, 10096–10103.
CrossRef - Gaussian 09, Revision E.01, Frisch, M. J.; Trucks, G. W.; Schlegel, H. B.; Scuseria, G. E.; Robb, M. A.; Cheeseman, J. R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G. A.; Nakatsuji, H.; Caricato, M.; Li, X.; Hratchian, H. P.; Izmaylov, A. F.; Bloino, J.; Zheng, G.; Sonnenberg, J. L.; Hada, M.; Ehara, M.; Toyota, K.; Fukuda, R.; Hasegawa, J.; Ishida, M.; Nakajima, T.; Honda, Y.; Kitao, O.; Nakai, H.; Vreven, T.; Montgomery, J. A., Jr.; Peralta, J. E.; Ogliaro, F.; Bearpark, M.; Heyd, J. J.; Brothers, E.; Kudin, K. N.; Staroverov, V. N.; Kobayashi, R.; Normand, J.; Raghavachari, K.; Rendell, A.; Burant, J. C.; Iyengar, S. S.; Tomasi, J.; Cossi, M.; Rega, N.; Millam, J. M.; Klene, M.; Knox, J. E.; Cross, J. B.; Bakken, V.; Adamo, C.; Jaramillo, J.; Gomperts, R.; Stratmann, R. E.; Yazyev, O.; Austin, A. J.; Cammi, R.; Pomelli, C.; Ochterski, J. W.; Martin, R. L.; Morokuma, K.; Zakrzewski, V. G.; Voth, G. A.; Salvador, P.; Dannenberg, J. J.; Dapprich, S.; Daniels, A. D.; Farkas, Ö.; Foresman, J. B.; Ortiz, J. V.; Cioslowski, J.; Fox, D. J. Gaussian, Inc., Wallingford CT, 2009.
- Rassolov, V. A.; Ratner, M. A.; Pople, J. A.; Redfern, P. C.; Curtiss, L. A., 2001, Journal of Computational Chemistry.
- Kiwiel, K. C. Mathematical Programming (Series A). 2001, 90 (1). Berlin, Heidelberg: Springer. 1–25
- Michael, S. L.; Freddie, R. S. J.; and Charles, L. B. The Journal of Chemical Physics 2002, 116,10606
CrossRef - Nguyena E, Bechet C,; Geuzaineb, L. N,; Computational Materials Science 2011 1-28
- Price, D. J.; Brooks, C. L. J. Chem. Phys., 2004, 121, 10096–10103.
CrossRef - Genheden, S.; Ryde, U. Expert Opinion on Drug Discovery. 2015 10(5) 449-461.
CrossRef - Verona, M. D.; Verdolino, V.; Palazzesi, F. and Corradini, R. Scientific Reports, 2017 7, 42799
CrossRef - Mall, V. S. and Tiwari, R. K. AIP Conference Proceedings 2018, 1953, 140118 https://doi.org/10.1063/1.5033293
CrossRef - 24. Mall, V. S.; Pandey, V. and Tiwari, R. K. International Journal of Innovative Research and Review 2018, 5(4), 59-63
- Ojha, R. P. and Tiwari, R. K. Journal of Biosciences 1999, 24 446.
- Agrawal, S,; Ojha, R. P.; Maiti, S. J. Phys. Chem. B 2008, 112, 6828–6836.
CrossRef - Wu J, Meng Q, Ren H, Wang H, Wu J, Wang Q. Acta Pharmacologica Sinica. 2017, 38(6), 798-805.
CrossRef - Gupta P, Muse O & Rozners E Biochemistry 2012 51, 63–73.
CrossRef - Gupta P, Zengeya T & Rozners E Chem Commun 2012 47, 11125–11127.
CrossRef - Riccirdi, A. S.; Quijano, E.; Putman, R.; Saltzman, W. M. and Glazer, P. M. Molecules 2018 23(3).

This work is licensed under a Creative Commons Attribution 4.0 International License.









