A Study for Transition of Vitamin C and Water Through Channel Membrane and Channel Aquaporin
Sogand Tohidi and Abdolhossein Rustaiyan
Department of Chemistry, Science and Research Branch, Islamic Azad University, Tehran, Iran.
Corresponding Author E-mail: m_monajjemi@yahoo.com
DOI : http://dx.doi.org/10.13005/ojc/340157
In this work we have exhibited vitamin C permeability and water transition through channel of Aquaporin. This study describes the investigation of the vitamin channel membrane proteins for selective transition among the B group vitamins and water. The interaction among the channels with some vitamins and water has been calculated. Total energy (Etot), Potential (Epot) and Kinetic (Ekin) energies (kcal/mol), calculated by the molecular simulation with different force fields in different temperatures. The NMR parameters have been calculated for confirming of the results.
KEYWORDS:Monte Carlo; NMR; Vitamin C; Channel Membrane; Aquaporin
Download this article as:| Copy the following to cite this article: Tohidi S, Rustaiyan A. A Study for Transition of Vitamin C and Water Through Channel Membrane and Channel Aquaporin. Orient J Chem 2018;34(1). |
| Copy the following to cite this URL: Tohidi S, Rustaiyan A. A Study for Transition of Vitamin C and Water Through Channel Membrane and Channel Aquaporin. Orient J Chem 2018;34(1). Available from: http://www.orientjchem.org/?p=41774 |
Introduction
Aquaporin or water channel, is a membrane proteins from intrinsic protein which form pores in the cell membranes, basically facilitating transition of water and some small molecules from outside to inside of a cell. The cell membrane of different animal and plants even bacteria and fungi, cell contain aquaporin through which water and some small molecules can flow more rapidly into and out of the cell via diffusing through phospholipid bilayers. The protein of aquaporin is composed of six transmembrane α-helices which are embedded in the cell membranes. The amino segment and carboxyl group ends face the inside of the cell and both of them halves resemble each other [1-5].
Vitamin C, or ascorbic acid, is a water-soluble compound which is naturally there is in some plants, foods, and available as a diet supplements. Vitamin C is needed for important bio synthesis of collagen and certain neurotransmitters and it is also involved in protein metabolism6.
Collagen is an important element, which plays a vital role in wound healing7. Vitamin C is also a unique physiological antioxidant8 and has been exhibited for regenerating other anti-oxidants within the human bodies, including vitamin E9.Furthermore to its antioxidant function, it plays a basic role in immune function4 and improves the absorption of iron for hemoglubin10.
Insufficient concentration of it in human body intake causes scurvy, which is characterized by lassitude5-9, widespread connective tissue10 weakness11, and capillary fragility5 11. Cells accumulate12 vitamin C through a 2th specific transports proteins and physiologic importance of de-hydro-ascorbic acids uptake to overall vitamin C is unknown12. The total body ranges of vitamin C are from 300 mg to about 2 g8-11 . High amount of vitamin C is maintained in cell and tissues11, and are highest in leukocyte, eye, adrenal gland, pituitary glands, and brains. Relatively low levels of vitamin C are found in extracellular fluid, such as plasma, red blood cell, and saliva11.
Model of Theory and Computational Details
Thermodynamic averages of molecular properties can be determined from molecular mechanics and Monte Carlo (MC) method, as can minimum-energy structure. Furthermore, it might be mention that constraining potential are occasionally applied at a finite temperature. MC simulations, which can generate canonical ensembles, are used when a system has difficult integral to be solved and have to generate some random numbers for generating uniform independent value statistically. In the MC, the metropolis algorithms are applied more than other algorithms because of its simplicities.
MC is the terms used in several fields of sciences and engineering or statistics and mathematics for meaning entirely different thing. The only one thing that all MC method has a same root is that those all needs random number for helping calculating something. MC simulation is widely used in the area of chemistries, biology, physic and applied engineering for ordering to determine the minimized structural and thermodynamic data of the systems at the atomic levels. Introducing the suitable potentials enable one for defining cluster of desired size. The MC is one of the important commonly used numerical techniques, with application in the statistical physics, quantum mechanics, and field theories and so on.
The accuracy of the method is defined by a random displacement. In this investigation, differences of force fields are shown by comparing the calculated energies using AMBER and OPLS. In this investigation Hyper-Chem professional release 7.01 is used for most of chemical calculations.
In this work, the quantum and molecular mechanic chemical studies were carried out using MC simulation. In the work, it has been investigated the temperature effects (291, 293, 295, 297, 298 and 300) for calculation of various aquaporin with OPLS, BIO+ and Amber force fields in several molecular mechanic (MM) method. The temperature range from 291 to 300 have selected based on the temperatures under and close to the normal temperature of the human body.
Different force fields are available in the MM program. Choosing the suitable force fields that is well parameterized for the molecular system under study is very important.
Gauge including atomic orbitals or GIAO and CSGT are adopted for solving the gauge problem in the calculation of nuclear magnetic shielding. The quantum chemical calculations yield the CS tensors in principal axes system which is: (σ33> σ22 > σ11)
The calculation was accomplished using Hyper-Chem professional release 7.01 package and the total energies, Potentials and Kinetic energies, have been done through MC simulation with AMBER, OPLS and MM+ in different Temperatures.
NMR chemical shifts measurements have been implemented in vast ranges of problems in chemistry and has unraveled to be an invaluable microscopic probes which have played a significant roles in the structural meaning.
Therefore, they used to evaluate the isotropic chemical-shielding or CSI, anisotropic chemical-shielding or CSA parameter, anti-symmetry parameter η.
CSI (ppm) = (σ33+ σ22 + σ11)/3
CSA (ppm) = σ33-(σ22 + σ11)/2
η= (σ22 – σ11)/( σ33 – CSI)
The Span factor according to the: Ω = σ33 – σ11. and κ=3(σiso –σyy ) / Ω
This work has been down based on previous theoretical and calculations13-40.
Results and Discussion
All data are shown in Figs.1-6 and tables 1-2.
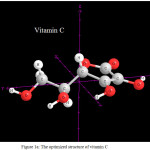 |
Figure 1a: The optimized structure of vitamin C Click here to View figure |
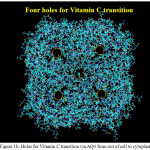 |
Figure 1b: Holes for Vitamin C transition via AQ4 from out of cell to cytoplasm Click here to View figure |
 |
Figure 2. 1: Hole in the Z-axis of AQ porin with 4OJ2 characterization .pdb file Channels Click here to View figure |
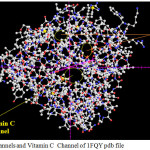 |
Figure 3: H2o Channels and Vitamin C Channel of 1FQY pdb file |
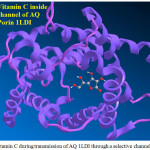 |
Figure 4: Vitamin C during transmission of AQ 1LDI through a selective channel Click here to View figure |
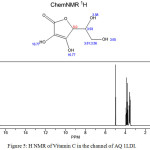 |
Figure 5: H NMR of Vitamin C in the channel of AQ 1LDI. |
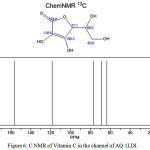 |
Figure 6: C NMR of Vitamin C in the channel of AQ 1LDI. |
Table 1: Gibbs free energy of various media with various force fields for AQP4+Vitamin C
| MM+ | AMBER | OPLS | ||||||
| Medium | Dielectric constant | ΔG(kcal/mol) | ||||||
| AQP1LDI +Vitamin C | Gas phase | 1 | 323.11 | 245.16 | 212.8 | |||
| Water | 78.2 | 399.48 | 316.23 | 348.44 | ||||
| Methanol | 32.1 | 429.18 | 421.06 | 369.7 | ||||
| Ethanol | 24.1 | 423.64 | 438.27 | 376.9 |
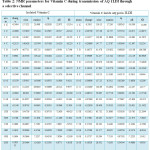 |
Table 2: NMR parameters for Vitamin C during transmission of AQ 1LDI through a selective channel Click here to View table |
NMR investigation gives deeper physical insight into the impact of different structures. In this work NMR parameters were calculated the aquaporin with different situation group in their active sites that the consider models geometry optimization were accomplished by method Monte Carlo. In fact, we calculate NMR factors and then compare data and we draw diagrams associated with them According to tables 1&2. The NMR measurements were carried out using GIAO method of nuclear magnetic resonance at theoretical concepts in different dielectric constants.
NMR calculations on trance membrane proteins via interaction with vitamin C using density functional theory (DFT) reveal that methods including electron correlation show significant improvements in the NMR shielding over results. Once of AQ 1LDI was shown to be a vitamin pore, studies began towards elucidating its structure, pore properties, and the mechanisms underlying selective water conduction and proton exclusion. In this work we have shown a significant increase in vitamin C permeability was observed in of AQ 1LDI as suitable protein trans-membranes for various vitamins resulting in dramatic swelling followed by dielectric of solvent tables1. Aquaporin, of AQ 1LDI, have been detected in different, non-overlapping areas of the central nervous system .This study describes the developmental expression pattern of the Vitamin C channel membrane proteins for some selected vitamin C. Total energy (E tot), Potential (E pot) and Kinetic (E kin) energy (kcal/mol), calculated by Monte Carlo simulation with Amber and OPLS force field in different Temperature the total energy, Potential and Kinetic energy (kcal/mol). The Total energy (E tot), Potential (E pot) and Kinetic (E kin) energy (kcal/mol), are calculated by Amber and OPLS force field. In this study, we examined Energy values from interaction of aquaporin with different vitamins in different media of solvents (water, methanol, ethanol and DMSO) in different temperature. Calculations of the Total energy, potential energy and kinetic energy by Monte Carlo simulation (AMBER, OPLS) have been to solvents (water, methanol, ethanol and DMSO) in different temperature and in Different number of solvents. Since the two different force fields have been utilized, the calculated energy of molecules will not be the same.
References
- Agre P. Proc Am Thorac Soc. 2006, 3 (1): 5–13
CrossRef
- Knepper MA, Nielsen S. J. Am. Soc. (2004). 15 (4) doi:10.1097/01. ASN.0000118814.47663. 7D.
CrossRef
- Schrier RW , Drug News Perspect. 2007, 20 (7): 447–53. doi:10.1358/dnp. 2007. 20.7.1138161.
- Parisi M, Dorr RA, Ozu M, Toriano R , J Biol Phys. 2007, 33 (5–6): 331–43. doi:10.1007/s10867-008-9064-5.
CrossRef
- Paganelli CV, Solomon AK. J. Gen. Physiol. 1957, 41 (2): 259–77. doi:10.1085/jgp.41.2.259.
CrossRef
- Li Y, Schellhorn HE. J Nutr 2007;137:2171-84.
CrossRef - Carr AC, Frei B. Am J Clin Nutr 1999, 69, 1086-107.
CrossRef - Frei B, England L, Ames BN. Proc Natl Acad Sci U S A 1989, 86, 6377-81.
CrossRef - Jacob RA, Sotoudeh G. Nutr Clin Care , 2002, 5:66-74.
CrossRef - Gershoff SN. , Nutr Rev , 1993, 51, 313-26.
CrossRef - Wang AH, Still C. Nutr Clin Pract 2007, 22, 445-8.
CrossRef - Stephen R, Utecht T. J Emerg Med 2001, 21, 235-7.
CrossRef - Monajjemi, M.; Najafpour, J. Fullerenes, Nanotubes, and Carbon Nanostructures, 2014, 22(6): 575–594.
CrossRef - Mahdavian, L.; Monajjemi, M.; Mangkorntong, N. Fullerenes, Nanotubes and Carbon Nanostructures, 2009, 17 (5), 484-495.
CrossRef - Monajjemi,M.; Nayyer T. Mohammadian J. Comput. Theor. Nanosci. 2015, 12, 4895-4914.
CrossRef - Monajjemi, M.; Lee, V.S.; Khaleghian, M.; B. Honarparvar, B.; F. Mollaamin, F. J. Phys.Chem C. 2010, 114, 15315.
CrossRef - Monajjemi, M. Struct Chem. 2012, 23,551–580.
CrossRef - Monajjemi, M.; Boggs, J.E. J. Phys. Chem. A, 2013, 117, 1670 −1684.
CrossRef - Monajjemi, M.; Khaleghian, M, Journal of Cluster Science. 2011, 22 (4),673-692.
CrossRef - Monajjemi, M.; Wayne Jr, Robert. Boggs, J.E. Chemical Physics. 2014, 433, 1-11.
- Monajjemi, M. Falahati, M.; Mollaamin, F.; Ionics, 2013, 19, 155–164.
CrossRef - Monajjemi, M.; Mollaamin, F. Journal of Cluster Science, 2012, 23(2), 259-272.
CrossRef - Tahan, A.; Monajjemi, M. Acta Biotheor, 2011, 59, 291–312.
CrossRef - Mollaamin, F.; Monajjemi, M. Physics and Chemistry of Liquids .2012, 50, 5, 2012, 596–604.
- Monajjemi, M.; Khosravi, M.; Honarparvar, B.; Mollaamin, F.; International Journal of Quantum Chemistry, 2011, 111, 2771–2777.
CrossRef - Monajjemi, M. Theor Chem Acc, 2015, 134:77 DOI 10.1007/s00214-015-1668-9.
CrossRef - Monajjemi, M. Journal of Molecular Modeling , 2014, 20, 2507.
CrossRef - Monajjemi, M.; Khaleghian, M.; Mollaamin, F. Molecular Simulation. 2010, 36, 11, 865.
- Monajjemi, M. Biophysical Chemistry. 2015 207,114 –127.
CrossRef - Jalilian,H.; Monajjemi, M. Japanese Journal of Applied Physics. 2015, 54, 8, 08510.
- Monajjemi, M.; Mollaamin, F. Journal of Computational and Theoretical Nanoscience, 2012, 9 (12) 2208-2214.
CrossRef - Mollaamin F, Varmaghani Z, Monajjemi M. 2011; 49: 318-336.
- Mollaamin, F.; Baei, MT.; Monajjemi, M.; Zhiani, R.; Honarparvar, B.;49. Russian Journal of Physical Chemistry A, Focus on Chemistry, 2008, 82 (13), 2354-2361
CrossRef - M Monajjemi, L Mahdavian, F Mollaamin, B Honarparvar,Fullerenes, Nanotubes and Carbon Nanostructures,2010, 18 (1), 45-55.
CrossRef - Monajjemi, M. Falahati, M.; Mollaamin, F.; Ionics, 2013, 19, 155–164.
CrossRef - Monajjemi, M., Chahkandi, B. Journal of Molecular Structure: THEOCHEM, 2005, 714 (1), 28, 43.
- M Monajjemi, L Mahdavian, F Mollaamin, B Honarparvar, Fullerenes, Nanotubes and Carbon Nanostructures,2010, 18 (1), 45-55
CrossRef - Tian Lu, Feiwu Chen, Sinica, 69, 2011, 2393-2406.
- Tian Lu, Feiwu Chen, J. Mol. Graph. Model, 2012, 38, 314-323.
CrossRef - Tian Lu, Feiwu Chen, Multiwfn: A Multifunctional Wave-function Analyzer, J. Comp. Chem. 2012, 33, 580-592.
CrossRef

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.

This work is licensed under a Creative Commons Attribution 4.0 International License.









