Synthesis, Structure-Activity Relationship, Docking and Molecular Dynamic Simulation of Curcumin Analogues Against HL-60 for Anti Cancer Agents (Leukemia)
Yum Eryanti1, Adel Zamri1, Neni Frimayanti2, Hilwan Yuda Teruna1, Unang Supratmman3, Tati Herlina3 and Yoshihito Shiono4
1Department of Chemistry, Faculty of Mathematics and Natural Sciences Universitas Riau Kampus Bina Widya, Jl. HR Subantas KM 12.5 Pekanbaru 28293, Indonesia.
2Sekolah Tinggi Ilmu Farmasi (STIFAR) Riau, Jalan Kamboja, Pekanbaru 28293, Indonesia.
3Department of Chemistry, Faculty of Mathematics and Natural Sciences, Padjadjaran University, Jalan Raya Bandung-Sumedang Km 21, Jatinangor 45363, Sumedang, Indonesia.
4Department of Bioresource Engineering, Faculty of Agriculture, Yamagata University, 1-23 Wakabamachi, Tsuruoka 997-8555, Japan.
Corresponding Author E-mail: adel.zamri@lecturer.unri.ac.id
DOI : http://dx.doi.org/10.13005/ojc/330503
Cancer such as leukemia is one of the dangerous diseases in the world. New agents with high activities are needed to attack this cancer. In this research, it was divided into three stages: synthesis, biological assay and computational approach such as molecular docking, molecular dynamic and structure activity (SAR) studies to ensure the biological activity. Synthesis of 45 curcumin analogues has been done using base or acid catalyzed aldol condensation reaction. The biological activities human promyelocytic leukemia cells line (HL-60) of those 45 curcumin analogues were investigated using 48-hour continuous exposure MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay technique and Genetic algorithm technique in Autodock 4.0 was used for molecular docking. Based on biological assay results, eight of curcumin analogues (i.e. compound 5, 8, 11, 14, 17, 28, 34 and 41) are found to be active against HL-60. From the molecular docking studies, the results showed that eight of these curcumin analogues were highly potent against HL-60 for anti cancer. Finally, docking studies and MD showed that the presence of the hydroxyl group in the meta position can enhance the biological activity of compounds.
KEYWORDS:Curcumin; Docking; Leukemia; HL-60; MD Simulation
Download this article as:| Copy the following to cite this article: Eryanti Y, Zamri A, Frimayanti N, Teruna H. Y, Supratmman U, Herlina T, Shiono Y. Synthesis, Structure-Activity Relationship, Docking and Molecular Dynamic Simulation of Curcumin Analogues Against HL-60 for Anti Cancer Agents (Leukemia). Orient J Chem 2017;33(5). |
| Copy the following to cite this URL: Eryanti Y, Zamri A, Frimayanti N, Teruna H. Y, Supratmman U, Herlina T, Shiono Y. Synthesis, Structure-Activity Relationship, Docking and Molecular Dynamic Simulation of Curcumin Analogues Against HL-60 for Anti Cancer Agents (Leukemia). Orient J Chem 2017;33(5). Available from: http://www.orientjchem.org/?p=39020 |
Introduction
Cancer is a dangerous disease and one of the leading causes death in the world. Currently, the major treatments for cancer include surgery, chemotherapy, and radiation [1]. However, high incidences of undesirable side effects have prompted researchers to search for safer and more effective anti cancer agents with reduced side effects.
Curcumin is one of the most potent and multi-targeting phytochemicals against variety of cancers. The cancer preventive capability of curcumin is linked to its direct antioxidant ability to eliminate free radicals and to reduce oxidative stress [2]. In the last decade, a number of investigation have been conducted on synthetic modifications of curcumin, especially, are considered promising compounds and are widely reported to exert good antioxidant and antitumor activities. For example, monocarbonyl 5-carbon spacer, curcumin analogues having cycloalkanone or piperidone central motif (B) were found to have good antitumor and also antioxidant activities [3, 4]. In addition, curcumin have also been tested for its action in acute myeloblastic leukemia cell line HL-60. The results clearly show that curcumin induces apoptosis in these cells. Telomerase, a reverse transcriptase has been found in more than 80% of human cancers and, therefore, can be considered as a potential marker for tumorigenic [5, 6].
In this study, 45 curcumin analogues have been synthesized in completed series with added variation of functional group in ortho, meta and para positions. Thus far, there is no report about synthesized of curcumin analogue similar to this variation position. Moreover, they reported synthesis of curcumin analogues only with adding variation functional group [4-7]. The biological activity of these 45 curcumin analogues was then investigated by using 48-hour continuous exposure MTT assay technique. There are eight compounds i.e. compound 5, compound 8, compound 11, compound 14, compound 17, compound 28, compound 34 and compound 41 are found to possess good activity against HL-60. Molecular docking was applied for those eight compounds to ensure their activity. From the spatial arrangement, contributions of the ligand with the residues of the active site were determined.
Experimental
General Procedure for the Synthesis of Chalcones
Melting points were determined on an electro-thermal melting point apparatus and are uncorrected. The ultra violet (UV) spectra were obtained on a UV Ultraspec 3000 Pro spectrophotometer. The IR spectra were recorded on a Perkin-Elmer 1760X FT-IR (Waltham, MA, USA) in KBr. The mass spectra were recorded with a JEOL JMS-700 (Tokyo, Japan) and a SynaptG2 mass spectrometer (Waters, Milford, MA, USA). 1H and 13C-NMR spectra were recorded with an Agilent DD2 system (Santa Clara, CA, USA) operating at 500 (1H) and 125 (13C) MHz, using residual (δH 7.26) and deuterated solvent (δC 77.0) peaks of CDCl3 as reference. For synthesis procedure Mas-II Sineo Microwave was used. Chromatographic separations were carried out on silica gel 60 (Merck, Darmstadt, Germany). Thin layer chromatography (TLC) plates were precoated with silica GF254 (Merck, 0.25 mm) and detection was achieved by spraying with 10% H2SO4 in ethanol, followed by heating [8, 9].
General Procedure for Preparation of Curcumin
Some 45 monoketone curcumin analogues (Table 4) have been synthesized using base or acid catalyzed aldol condensation reaction of the appropriate substituted benzaldehyde and corresponding NH-4-piperidones, N-methyl-4-piperidones and N-benzyl-4-piperidones with a combinatorial chemistry approach. The reaction proceeded under microwave irradiation with reaction times 10 min at 300 Watt of power and temperature of 60oC to afford the products in good yields. The crude compounds were chromatographed on silica gel and further readily purified by re-crystallization.
A total of 0.01 mol piperidone associated and related aryl aldehyde 0.02 mol, was placed in a two-neck round bottom flask which was equipped with a magnetic stirrer and condenser, some 0.7 ml of 40% sodium hydroxide and 10 ml of absolute ethanol were added. The reaction mixture was treated as compound 1 and produced related compounds. Furthermore, the purity of the compounds was then analyzed using TLC, HPLC and melting point determination [10].
Biological Assay
The biological activity (cytotoxic on HL-60) of 45 curcumin analogues compounds were investigated using MTT assay. Cells HL-60 in the plates 96-well with cells density of 3 x 104 cells cm-3 were incubated for 24 h for cell growth. Then, it was added to the samples with various concentrations that have been dissolved in DMSO (dimethyl sulfide), this were done with a pH of 7.30 to 7.65 by addition of PBS (phosphate buffer solution), with DMSO solution as a control. Tests performed after incubated for 48 h, then reagent MTT (3- (4,5-dimetiltiazol-2-yl) -2,5-dipenil tetrazoliumbromida) was added and incubation was continued for 4 h. MTT reagent stop (consist of containing SDS (sodium dodecyl sulphate) was then added. Furthermore, it was incubated for 24 h. Optical density was read using a micro plate reader at the wave length (λ) of 570 nm.
Molecular Docking
Eight ligand and macromolecule (download from PDB data base www.pdb.org: PDB ID 3DU6) were prepared before the docking process. For the macromolecule was added polar hydrogen atoms and its non-polar hydrogen atoms were merged. Kollman charges were assigned and solvation parameters were added to this enzyme molecule. While, for the ligands, non-polar hydrogen atoms were merged with Gasteiger charges assigned. All rotatable bonds of ligands were set to be rotatable with 0.375 Å grids spacing around the catalytic triad. Docking was performed through Genetic Algorithm using Autodock 4.0 software package (http://autodock.scripps.edu/) with a grid box 55, 17, 42 dimensions along x, y, z axes. Upon the completion of the docking process, confirmation with the lowest energy was chosen. The hydrogen bonding, van der Waals and any other interactions were then analyzed using Discovery studio visualizer 3.0 (Accelrys) (http://accelrys.com/customer-support/contact.html) for better insight.
Molecular Dynamic Simulation
MD simulations for the modeled protein were performed using the program NAMD (Nanoscale Molecular Dynamics program; v 2.9) [11] and all files were generated using visual molecular dynamics (VMD) [12]. The protein was solvated with a TIP3P water box with a 2.5 Å layer of water for each direction of the coordinate structure and a CHARMM (Chemistry at HARvard Macromolecular Mechanics) was used as a force field.
Results and Discussion
Biological Assay
The biological activities against human promyelocytic leukemia cells line HL-60 of 45 synthesized curcumin analogues were investigated. Table 1 is presented the percentage of the viability 45 curcumin analogues at 0.5 μM and the curve of the viability 45 curcumin is presented in Figure 1. Base on Figure 1 red block is indicated low viability for then used to determine the IC50 value. Eight of curcumin analogues are found as active compounds against HL-60. The IC50 (µM) value for active curcumin analogues is presented in Table 2.
Table 1: The percentage of the viability 45 curcumin analogue at 0.5 μM.
|
compound |
Viability (%) |
compound |
Viability (%)) |
compound |
Viability (%) |
|
1 |
96,12 |
4 |
89,60 |
7 |
100,91 |
|
2 |
87,34 |
5 |
31,67 |
8 |
43,37 |
|
3 |
100,08 |
6 |
92,47 |
9 |
99,15 |
|
10 |
101,83 |
13 |
101,50 |
16 |
101,55 |
|
11 |
17,16 |
14 |
11,58 |
17 |
16,45 |
|
12 |
97,80 |
15 |
96,06 |
18 |
97,55 |
|
19 |
79,83 |
22 |
97,79 |
25 |
95,88 |
|
20 |
95,68 |
23 |
70,54 |
26 |
90,42 |
|
21 |
95,70 |
24 |
96,45 |
27 |
95,67 |
|
28 |
48,73 |
29 |
86,90 |
30 |
95,14 |
|
31 |
77,01 |
32 |
92,88 |
33 |
98,67 |
|
34 |
25,73 |
35 |
68,00 |
36 |
93,74 |
|
37 |
88,05 |
38 |
89,78 |
39 |
98,07 |
|
40 |
90,75 |
41 |
10,50 |
42 |
98,92 |
|
43 |
100,99 |
44 |
100,05 |
45 |
11,55 |
The range value of IC50 for eight of synthesized curcumin analogue is between 0.24-0.47 µM and curcumin has higher IC50 of 5.40. Based on Table 2, it seems that compound 11 give the best activity against HL-60, followed with compound 17, compound 14, compound 8, compound 34, compound 5, compound 28¸ and compound 41, indicated that compounds bearing hydroxyl group in the meta positions have better biological activities against HL-60, suggesting that they are promising as inhibitors for HL-60.
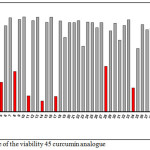 |
Figure 1: Curve of the viability 45 curcumin analogue Click here to View figure |
Table 2: IC50 value of human promyelocytic leukemia cell line (HL-60)
|
Compound |
IC50 (µM) |
Compound |
IC50 (µM) |
compound |
IC50 (µM) |
|
1 |
– |
4 |
– |
7 |
– |
|
2 |
– |
5 |
0,43 |
8 |
0,38 |
|
3 |
– |
6 |
– |
9 |
– |
|
10 |
– |
13 |
– |
16 |
– |
|
11 |
0,36 |
14 |
0,28 |
17 |
0,37 |
|
12 |
– |
15 |
– |
18 |
– |
|
19 |
– |
22 |
– |
25 |
– |
|
20 |
– |
23 |
– |
26 |
– |
|
21 |
– |
24 |
– |
27 |
– |
|
28 |
0,47 |
29 |
– |
30 |
– |
|
Compound |
IC50 (µM) |
Compound |
IC50 (µM) |
compound |
IC50 (µM) |
|
31 |
– |
32 |
– |
33 |
– |
|
34 |
0,42 |
35 |
– |
36 |
– |
|
37 |
– |
38 |
– |
39 |
– |
|
40 |
– |
41 |
0,24 |
42 |
– |
|
43 |
– |
44 |
– |
45 |
– |
|
curcumin |
5,40 |
Molecular Docking
The focus of molecular docking is to computationally stimulate the molecular recognition process while the aim of molecular docking is to achieve an optimized conformation for both protein and ligand. Docking play an important role in the rational design of drug design where it is frequently use to predict the preferred orientation and binding site of small molecule to their target protein in order to ascertain its affinity and activity.
In silico docking studies were performed to evaluate the effects of curcumin analogue against HL-60. From the docking results, compounds 5 showed two hydrogen bonds (green dashed line) between ligand and residues Lys515. Likewise, in the case of compound 8, only one hydrogen bond was observed between the ligand and the residue Lys515. Bayomi and co-workers reported that the active site on this protein is the protein have an interaction with the residue Lys169 [3] but it is not clear what kind of interaction. In our case, hydrogen bonding is formed with the residue Lys515. It is indicating that another possible mode of interaction between the ligand and this protein. The binding mode of these two ligands with enzyme is presented in Figure 2.
For compound 11, docking result indicated there are two hydrogen bonds were observed between hydroxyl group of ligand and backbone –NH of residue Lys515, and also between hydroxyl group of ligand and residue Asp440. It is assumed that hydroxyl group has striking effect for cytotoxic due to electronic and conjugated effect of electronic donating group. In addition, from the best docking pose of compound 11, residues
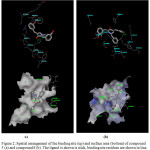 |
Figure 2: Spatial arrangement of the binding site (up) and surface area (bottom) of compound 5 (a) and compound 8 (b). The ligand is shown is stick, binding site residues are shown in line. Click here to View figure |
Arg 511 shown interaction with ligand through van der Waals interaction (grey area) suggesting the importance of this residue in the formation of van der Waals pocket.
The other curcumin analogue is compound no 14, similar to the previous ligand, this ligand was also docked. The spatial arrangement of the binding site indicated several residues playing important role in determining the binding interaction for the ligand. From the docking results, compound 14 were observed to exhibit hydrogen bonding interactions between the hydroxyl group of the ligand and the residue Asp440 and also between hydroxyl group of Tyr519 and the ligand. The binding site of compound 11, compound 14 with their surface area are presented in Figure 3.
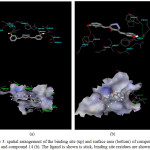 |
Figure 3: spatial arrangement of the binding site (up) and surface area (bottom) of compound 11 (a) and compound 14 (b). The ligand is shown is stick, binding site residues are shown in line. Click here to View figure |
The other active curcumin analogues are compound 28, compound 34 and compound 41. Similar to the previous ligand these ligands were also docked. From the docking results, compound 34 was observed the hydrogen bonding between the ligand and the residue Lys515 but the hydrogen bond interaction was not established for compound 28 and compound 41.
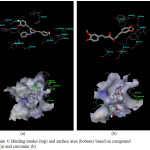 |
Figure 4: Binding modes (top) and surface area (bottom) based on compound 17 (a) and curcumin (b). Click here to View figure |
Compound 17 was docked into the enzyme. The best docking pose of compound 17 exhibited van der Waals interaction between the ligand and residue Arg511, suggesting the importance of this residue in the formation of the van der Waals specificity pocket. Compound 17 was also found to interact via hydrogen bonding of the hydroxyl group of the ligand and residue Lys515, between hydroxyl group of the ligand and Asp440. The higher number of hydrogen bond may account for ligand is more active. Figure 4 is depicted the binding modes for compound 17. As the other analogue compounds, curcumin as a parent compound were also docked into the enzyme. The spatial arrangement of the binding site indicated that there is no hydrogen bonding is established between the ligand and the enzyme. The van der Waals interaction and any other interaction were also not established. The binding mode of the curcumin and the enzyme is presented in Figure 4 More active compound will have more hydrogen bond interaction or van der Waals interaction [13]. In this case, base on docking results, curcumin is confirmed as not active compound against HL-60.
Thus, based on the docking results, we can propose the structure-activity relationship as mentioned below point by point: firstly, presence of hydroxyl group in meta position on ring A or ring C was found to enhance the biological activity of curcumin analogues by building the hydrogen bond with the residue such as Lys115, Asp440, and Tyr519. The general structure of curcumin is depicted in Figure 5. Secondly, presence of hydrogen, benzyl or methoxy in R” position on ring B. There was no specific effect to the biological activity of curcumin analogue. Thirdly, presence of methoxy group in ortho, meta, and para position on ring A or ring C was found to give better biological activity of curcumin analogues.
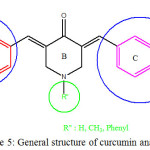 |
Figure 5: General structure of curcumin analogue Click here to View figure |
Molecular Dynamic
MD simulation was carried out on these eight compounds to further explore the ligand-receptor interaction [14]. In order to evaluate the stability of the MD simulation, the properties (i.e. H-bond) of each complex was inspected. Hydrogen bonding interaction is quite important in the binding between ligand and receptor. In this study, the MD simulations were performed at temperature 300 K to see the affinity of the ligand to the binding site. In general, the conformations of these seven ligands are maintained to binding with the important residues as presented in Figure 6 and Table 3.
Table 3: Interaction of ligand with amino acid
|
Ligand |
MD 300K |
Docking results |
|
Cpd5 |
Asp440, Phe441, Pro442, Lys514, Lys515, Tyr519 | Asp440, Phe441, Pro442, Lys514, Lys515 |
|
Cpd8 |
Phe441, Pro442, Cys445, Phe448, Lys514, Lys515 | Phe441, Pro442, Cys445, Phe448, Lys450, Arg511, Ala512, Lys514, Lys515 |
|
Cpd11 |
Phe441, Pro442, Arg511, Lys515 | Asp440, Phe441, Pro442, Arg511, Lys515 |
|
Cpd14 |
Phe441, Pro442, Arg511, Lys515 | Asp440, Phe441, Pro442, Lys515,Tyr519 |
|
Cpd17 |
Asp440, Phe441, Pro442, Arg511, Lys515 | Asp440, Phe441, Pro442, Arg511, Lys515, Tyr519 |
|
Cpd28 |
Phe441, Sys445, Lys515 | Phe441, Pro442, Cys445, Arg511, Lys514, Lys515 |
|
Cpd34 |
Asp440, Pro442, Cys445,Lys514 | Phe441, Pro442, Cys445, Lys514, Lys515 |
|
Cpd41 |
Phe441, Pro442, Lys515 | Phe441, Pro442, Lys514, Lys515 |
Based on Table 3, it can be observed that eight ligands during the docking process have hydrogen bond with the important residues such as: Asp440, Arg511, Lys515, and Tyr519. For MID simulation, at the temperature 300 K show that at the end of the simulation, these seven ligands seem lost their ability but still has interaction via hydrogen bonding with some of the important residues such as Asp440 and Lys515.
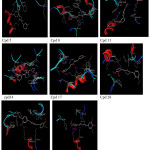 |
Figure 6: Visualization of the binding mode Click here to View figure |
Conclusion
Eight of curcumin analogues have been synthesized and they have good activity against HL-60 according to the biological assay. Docking and MD simulation studies were performed to evaluate the effects of curcumin analogue against HL-60. Docking studies showed that higher numbers of hydrogen bonding which validated them as potential candidates for second generation drug discovery. Bioassay studies of all curcumin analogues indicated that compounds such as compounds 11 and 17 bearing hydroxyl group in the meta positions have better biological activities against HL-60, suggesting that they are promising as potential inhibitors for HL-60.
Acknowledgements
We thank DIKTI Kementerian Pendidikan Nasional for the financial support through grant Hibah 2011-2013 and hibah kompetensi 2017.
References
- Gerard L. C. C.; Halimah, Y. Second report of national cancer register; cancer incidence in Malaysia, 2003, Kuala Lumpur, Malaysia.
- Anand, P.; Kunnumakkara A. B.; Newman R. A.; and Agarwai B. B. Bioavailability of curcumin: problems and promises. Mol Pharm. 2007, 4, 807-818.
CrossRef - Bayomi, S. M.; El-Kashef, H. A.; El-Ashmawy, M. B.; Nasr, M. N. A.; El-Sherbeny, M. A.; Badria, M. A.; Abou-Zeid, L. A.; Ghaly, M. A; Abdel Aziz, N. I. Synthesis and biological evaluation of new curcumin derivatives as antioxidant and antitumor agents. Med Chem Res. 2013, 22, 1147-1162.
CrossRef - Joseph, A.; Shah, C. C.; Kumar, S. S.; Alex, A. T.; Maliyakkal, N.; Moorkoth, S.; Mathew, J. E. Synthesis in vitro anticancer and antioxidant activity of thiadiazole substituted thiazolidin-4-ones. Acta Pharm. 2013, 63, 397-408.
CrossRef - Liu, Q.; Yong-B, P.; Ping, Z.; Lian-Wen, Q.; Mu, Z.; Ning, G.; E-Hu, L.; and Ping, L. 6-Shogaol induces apoptosis in human leukemia cells through a process involving caspase-mediated cleavage of eIF2α.. Molecular Cancer, (2013), 12,135-147.
CrossRef - Yousef, K. M.; El-Sherbeny, M. A.; S.El-Shafie, F.; Farag, H. A.; Al-Deeb, O. A.; Awadalla, S. A. Synthesis of Cuurcumin analogues as potential antioxidant cancer chemopreventive agents. Arch Pharm Weinh. 2004, 37, 42-54.
CrossRef - Shoude, Z.; Xia, L; Jigang, Z; Rongcai, Y.; Yanyan, D.; Honglin, L.; Hui, W.; Weidong, Z; Jacarelhyperol. An induced apoptosis in leukemia cancer cell through inhibition the activity of Bcl-2 proteins. BMC Cancer. 2014, 14, 689-700.
CrossRef - Eryanti, Y.; Herlina, T; Zamri, A; Shiono, Y; Awang, K.; Nadiah, S; Supratman, U. N-Benzyl-(3E,5E)-3,5-bis(2-hydroxybenzylidene)-4-piperidone. Molbank, 2015, M852, doi:10.3390/M852.
CrossRef - Eryanti, Y.; Herlina, T.; Zamri, A.; Shiono, Y.; Awang, K.; Nadiah, S.; Supratman, U. 3,5-Bis(2-hydroxybenzylidene)piperidin-4-one. Molbank, 2014 M825, doi:10.3390/M825.
CrossRef - Aditama, R.; Eryanti, Y.; Mujahiddin, D.; Shah, Y. M.; Hertadi, R. Determination of activities of human carbonic anhydrase II inhibitors from curcumin analogs. Tropical Journal of Pharmaceutical Research. 2017, 16 (4): 849-854.
CrossRef - Nelson, M. T.; Humphrey, W.; Gursoy, A. NAMD: a parallel, object-oriented molecular dynamics program. Int J Supercomput Appl, 1996, 10(4), 251–268.
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: visual molecular dynamics. J. Mol Graph. 1996, 14(1), 33–38. 27–28.
- Marzieh, Y.; Neni, F.; Chin Fei, C.; Kusaira, K.; Belal, O. N.; Sharifuddin, M. Z.; Zanariah, A.; Habibah, A. W.; Noorsaadah, A. R. QSAR, In silico and in vitro evaluation of chalcone derivatives as potential inhibitors for H1N1 virus neuraminidase. Med Chem Res. 2016, 1-10,doi:10.1007/s00044-016-1636-5.
CrossRef - Huiding, X.; Yupeng, Y.; Fang, X.;. Xiaoguang, Q.; Kaixiong, F.; Jijun. An Investigation of Molecular Docking and Molecular Dynamic Simulation on Imidazopyridines as B-Raf Kinase Inhibitors. Int. J. Mol. Sci. 2015, 16, 27350–27361.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.









