Comparative Modeling Approach upon Synthesizing Silver Nanoparticles from Solanum Virginianum
Balachandar Baskaran1, Vandana Sridhar2, Abinandan Sudharsanam3 and Praveen Kuppan4
1Madha Engineering College, Kundrathur, Chennai 600069, Tamilnadu, India.
2National Centre for Nanoscience and Nanotechnology, University of Madras, Chennai, India.
3EcoTech Labs Pvt Ltd, Ekkatuthangal, Chennai 600078, Tamilnadu, India.
4Ramky Enviro Engineers, Hyderabad, Telangana, India.
Corresponding Author E-mail: balachandarbt5@gmail.com
DOI : http://dx.doi.org/10.13005/ojc/330525
The major objective focusses on synthesizing silver nanoparticles (Ag-NPs) from Solanum virginianum under optimum conditions. In this study, mathematical tools such as central composite design (CCD) and artificial neural networking (ANN) used for identifying process optimizing parameters. The maximum wavelength recorded (λ=425 nm) using chemical mediated synthesis for the leaf extract. Based on the results, that plant extract concentration (15 g ml-1), temperature (65°C), time (11 mins) with pH (13) yielded the highest concentration 2mg g-1 of Ag-NPs. Furthermore, the statistical analysis yielded regression coefficient (R2) showing 0.96 indicating the RSM model & ANN model is in similar with the obtained investigational results. Interaction effect shows that plant extract concentration, time and pH were significant with p<0.05.
KEYWORDS:Solanum Virginianum; AgNPs; RSM; ANN
Download this article as:| Copy the following to cite this article: Baskaran B, Sridhar V, Sudharsanam A, Kuppan P. Comparative Modeling Approach upon Synthesizing Silver Nanoparticles from Solanum Virginianum. Orient J Chem 2017;33(5). |
| Copy the following to cite this URL: Baskaran B, Sridhar V, Sudharsanam A, Kuppan P. Comparative Modeling Approach upon Synthesizing Silver Nanoparticles from Solanum Virginianum. Orient J Chem 2017;33(5). Available from: http://www.orientjchem.org/?p=38894 |
Introduction
Advanced material such as metal derived nanoparticles have extensive applications due to its unique surface properties [1-4]. There are various techniques for producing silver nanoparticles viz., chemical route, photoreduction in reverse micelles [5-10]. Since nanoparticles are used widely for applications that necessitates for emergent an eco-friendly and cost-effective process [11]. The biological sources (microbial and plant biomass) could be a substitute approaches for nanoparticles synthesis in an eco-friendly manner [12]. However, plant route is more advantageous due to the painstaking maintenance of microbial cultures. [13]. Hence plant route attracted the scientific community to identify potential sources and its applications. For instance, plant sources such as Alfalfa, Aloe vera, Cinnamon camphora, Carica sp, Parthenium hysterphorrus, Diopyrus kaki, Hibiscus rosasinensis, have shown potential for the synthesis of nanoparticles is yet to be fully explored [14-17].
Here we have used the leaf extract of Solanum virginianum to synthesize silver nanoparticles. These prove to be both as reducing agent and capping agent. Solanum virginianum (family: solanaceae) is an important medicinal weed in traditional ayurvedic health care systems. The root extract is used to cure urinary diseases, cough and hair fall. Leaf extract is useful in the treatment of fever, cough, Asthma, Sore throat, Rheumatism and treated for gonorrhea, influenza and enteric fever [18]. Optimization of process parameters helps the production of high-quality nanoparticles minimizing the variable interventions. Response surface methodology is one such tool to used various design experiments which render in less time consuming rather than usual laborious trials. The application of central composite design (CCD) is useful to study the interactions among variables and optimize the experimental conditions based on second order polynomial equation. Furthermore, these designs enhance the process settings against other influencing factors [19, 20]. In this work, CCD used for optimization of process parameters like reaction time, temperature, pH and concentration of plant extract which affects the bioreduction of silver nitrate. Furthermore, the application of ANN modeling has also been used to validate the experimental data.
Materials And Methods
Plant Material and Synthesis of Silver Nanoparticles
Solanum virginianum leaves from the local orchard were taken adjacent the institute at Chennai, Tamilnadu, India. The leaves were washed, dried and prepared as an extract by boiling and filtering it. 0.01M silver nitrate supplemented to extract from the leaves to scale up the volume up to 25 ml and heated at various temperatures, pH condition. A variation in color was perceived during the heating process.
Spectra Investigation
The reduction of silver (Ag) ions from the absorption (UV-Vis) spectrum of the reaction mixture at various wavelength to ensure the maximum absorbance (as shown in fig 1) through spectrophotometer.
Standard Graph
The standard graph resulted by varying the nanoparticles concentration (3000-15000µg/ml) in millipore water. The graph measured as the volume of silver nanoparticle versus OD absorbance at 425nm.
Response Surface Optimization
RSM involved in the optimization of various parameters for silver nanoparticle synthesis by bioreduction method. CCD obtained the statistical method with four independent variables (pH, reaction time, temperature, the concentration of plant extract). Fractional factorial CCD generated 31 experiments based on three factors for five different levels. Upon completion of the experiments, the OD of nanoparticle production used as the output. The polynomial model obtained by regression techniques for four factors using MINITAB 17 determines the optimal response region of the silver nanoparticles yield from bioreduction of silver nitrate.
Z=λ0+λ1X1+ λ2X3+ λ3X3+ λ4X4+ λ11X12+ λ22X22+ λ33X32+ λ44X42+ λ12X1X2+ λ14X1X3+ λ14X1X4+ λ23X2X3 + λ24X2X4+ λ34X3X4 ………. (1)
Where Z is the output, X1–4 are the input variables, λ0 is the regression constant , λ1–4 the linear effects, λ11-44 the squared effects and λ12,13, 14, 23, 24, 34 are interaction terms.
ANN Modelling
The input of ANN is weighted, and the sum of the weighted inputs along with bias values will help to support the input values to next function (Vedaraman et al. 2017). This function will be able to determine the input/output behavior that also contributes to nonlinear modeling and stability of neural network involved in the process. These input values processed with neuron and the output will be developed based on the propagation model. In this study, a commonly used forward feed propagation model (Fig 1) was used to predict the concentration of silver nanoparticles. In the neural network, each neuron is embedded to another set of a neuron through adaptable weight. Levenberg Marquardt, the gradient descendent method was adopted as a model training and learning function.
Results and Discussion
UV-Vis Spectra Analysis and Standard Graph
A color change of reaction mixture because of the ionic reduction into Ag particles owing to surface plasmon resonance with plant extract. The result found paves the way for identifying potential weeds for synthesizing Ag nanoparticles. The standard graph shown in figure 1 was prepared by varying the nanoparticles concentration (3000-15000µg ml-1) in millipore water at 425nm. The absorbance values plotted against concentration to obtain regression equation.
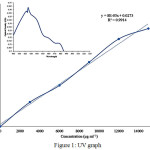 |
Figure 1: UV graph Click here to View figure |
Optimization of Silver Nanoparticle Production by RSM Method
Second order polynomial equation is used in CCD to generate the predicted data based on the input and variables (Table 1). Moreover, the surface plots under CCD helps to exemplify input based on single and interactive effects of the response. Also, the second order regression equation post-ANOVA analysis yields the quantity of Ag-NPs synthesized on assigned input of the variables (Table 2 & Table 3). Based on the coefficients of ANOVA the equation was fitted accordingly and are shown below
Z=0.2029 – 0.2529 X1 – 0.1540 X2 + 0.0426 X3 – 0.4658 X4 + 0.0912 X12 + 0.0678 X22 – 0.0035 X33 + 0.3314 X44 – 0.0997 X1*X2 + 0.0253 X1*X3 + 0.1557 X1*X4 + 0.0024 X2*X3 + 0.1346 X2*X4 – 0.0222 X3*X4
The response model shows that experimental data and predicted output are very linear with R2>0.9 (Table 1) close to unity indicating model can elucidate 96% response variability. Furthermore, the linear and quadratic terms based on the model (Table 2) were significant for plant extract, temperatureure, time and pH (p<0.05).
Table 1: Experimental and predicted results based on CCD and ANN
|
Std order |
Plant Extract (X1) |
Temperature (X2) |
Time (X3) |
pH (X4) |
Exp. data |
Predicted (RSM) |
Predicted (ANN) |
|
1 |
5 |
40 |
2 |
4 |
0.112 |
-0.02936 |
0.37247 |
|
2 |
25 |
40 |
2 |
4 |
0.196 |
0.313783 |
0.22491 |
|
3 |
5 |
90 |
2 |
4 |
0.148 |
0.2041 |
0.45883 |
|
4 |
25 |
90 |
2 |
4 |
0.317 |
0.148492 |
0.22949 |
|
5 |
5 |
40 |
20 |
4 |
0.164 |
0.044767 |
0.32101 |
|
6 |
25 |
40 |
20 |
4 |
0.226 |
0.489158 |
0.055398 |
|
7 |
5 |
90 |
20 |
4 |
0.201 |
0.287975 |
0.57601 |
|
8 |
25 |
90 |
20 |
4 |
0.519 |
0.333617 |
0.35221 |
|
9 |
5 |
40 |
2 |
10 |
0.16 |
0.366017 |
1.5165 |
|
10 |
25 |
40 |
2 |
10 |
1.419 |
1.331908 |
1.3087 |
|
11 |
5 |
90 |
2 |
10 |
1.401 |
1.137725 |
2.22 |
|
12 |
25 |
90 |
2 |
10 |
1.565 |
1.704867 |
0.25509 |
|
13 |
5 |
40 |
20 |
10 |
0.183 |
0.351392 |
1.287 |
|
14 |
25 |
40 |
20 |
10 |
1.454 |
1.418533 |
1.1437 |
|
15 |
5 |
90 |
20 |
10 |
1.23 |
1.13285 |
1.5744 |
|
16 |
25 |
90 |
20 |
10 |
1.66 |
1.801242 |
0.060269 |
|
17 |
-5 |
65 |
11 |
7 |
0 |
0.062025 |
0.97833 |
|
18 |
35 |
65 |
11 |
7 |
1.1561 |
1.073558 |
0.29779 |
|
19 |
15 |
15 |
11 |
7 |
0.342 |
0.166158 |
0.50779 |
|
20 |
15 |
115 |
11 |
7 |
0.627 |
0.782325 |
0.27668 |
|
21 |
15 |
65 |
0 |
7 |
0.023 |
0.103492 |
0.16938 |
|
22 |
15 |
65 |
29 |
7 |
0.375 |
0.273992 |
0.74805 |
|
23 |
15 |
65 |
11 |
1 |
0.5415 |
0.596992 |
2.4568 |
|
24 |
15 |
65 |
11 |
13 |
2.536 |
2.459992 |
0.19032 |
|
25 |
15 |
65 |
11 |
7 |
0.203 |
0.202918 |
0.19032 |
|
26 |
15 |
65 |
11 |
7 |
0.202429 |
0.202918 |
0.19032 |
|
27 |
15 |
65 |
11 |
7 |
0.203 |
0.202918 |
0.19032 |
|
28 |
15 |
65 |
11 |
7 |
0.203 |
0.202918 |
0.19032 |
|
29 |
15 |
65 |
11 |
7 |
0.203 |
0.202918 |
0.19032 |
|
30 |
15 |
65 |
11 |
7 |
0.203 |
0.202918 |
0.19032 |
|
31 |
15 |
65 |
11 |
7 |
0.203 |
0.202918 |
0.19032 |
Table 2: Estimated regression coefficient of second order polynomial model
| Coefficient | Estimated coefficient | t-value | p-value |
| λ0 | 0.2029 | 3.10 | 0.007 |
| λ1 | 0.2529 | 7.15 | 0.000 |
| λ2 | 0.1540 | 4.36 | 0.000 |
| λ3 | 0.0426 | 1.21 | 0.245 |
| λ4 | 0.4658 | 13.17 | 0.000 |
| λ11 | 0.0912 | 2.82 | 0.012 |
| λ22 | 0.0678 | 2.09 | 0.053 |
| λ33 | -0.0035 | -0.11 | 0.914 |
| λ44 | 0.3314 | 10.23 | 0.000 |
| λ12 | -0.0997 | -2.30 | 0.035 |
| λ13 | 0.0253 | 0.58 | 0.567 |
| λ14 | 0.1557 | 3.60 | 0.002 |
| λ23 | 0.0024 | 0.06 | 0.956 |
| λ24 | 0.1346 | 3.11 | 0.007 |
| λ34 | -0.0222 | -0.51 | 0.615 |
Table 3: Analysis of variance (ANOVA) for optimization of silver nanoparticles (R2=96%)
| Source | Degrees of freedom | sum of squares | Mean square | F-value | P-value |
| Regression | 14 | 11.5252 | 0.82323 | 27.44 | 0.000 |
| Linear | 4 | 7.3541 | 1.83851 | 61.29 | 0.000 |
| Square | 4 | 3.3164 | 0.82910 | 27.64 | 0.000 |
| Interaction(Two-way) | 6 | 0.8548 | 0.14246 | 4.75 | 0.006 |
| Residual Error | 16 | 0.4799 | 0.03000 | ||
| Total | 30 | 12.0051 |
The response surface plots relating combined effects among variables for silver nanoparticles synthesis depicted in figure 2 resulting from two variables constant at their middle level. Fig.2a shows the interaction outcome of plant extract (X1), pH (X4) on nano particle’s yield. It was observed that at initial levels of pH, the synthesis of nanoparticles was very low even there is an increase in plant extract concentration. Although, when the pH increases along with higher extract showed a good increase in the AgNPs synthesis. This may be because higher pH supported the stability of nanoparticles. These results were consistent with the literature (Singh et al., 2009; Iravani et al., 2014).The effect of temperature (X2) and plant extract (X1) on the AgNPs synthesis is shown in Fig 2b.It can be noted that lower levels of plant extract reflected in higher absorbance with an increase in temperature. However, a higher concentration of AgNPs observed at lower temperature levels with an increase in plant extract concentration. This may attribute to the increase in response proportions owing to temperature deviations, and the result is inconsistent with literature (Jiang et al., 2011).However, Sun et al 2014, in his study observed that effect of temperatures doesn’t contribute as the nanoparticles have reached the maximum level of extraction from plant source. The increase in pH with temperature showed a positive effect in the AgNPs synthesis (Fig 2c).
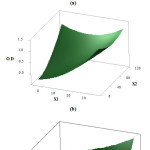 |
Figure 2 Click here to View figure |
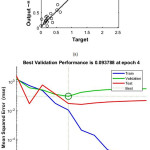 |
Figure 3 Click here to View figure |
The application of ANN modeling to the experimental data under Tansig and LM algorithm was used. To develop network, the data were separated into training (70%), validation (15%) and testing (15%) based on Levenberg-Marquardt calculations. From the fig 3a & b after five epoch with the value of 0.09378 and R2 value close to 1 indicating the goodness of the model. Furthermore, the parameters such pH, temperature and plant extract were significant with P<0.05 compared to other interaction factors.
Conclusion
The extraction of AgNPs was extracted using chemical mediated methods based on central composite design. It was observed that higher pH supported the increased extraction of AgNPs from the solution. This was significant with other two parameters such as temperature and plant extract with p<0.005.Both the model viz., RSM & ANN showed regression coefficient of 0.96 which is very close to 1 indicating the data’s are statistically significant. This study demonstrates that plant extract would be viable source for nanoparticles synthesis compared to other sources which is of many significant applications.
References
- Catauro, M.; Raucci, M.G.; De Gaetano, F.; Marotta, A. J. Mat. Sci. Mat. Med. 2004, 15, 831-837.
CrossRef - Crabtree, J.H.; Burchette, R.J.; Siddiqi, R.A.; Huen, I.T.; Hadnott, L.L.; Fishman, A. Peri Dia. Int. 2003, 23,368-74.
- Królikowska, A.; Kudelski, A.; Michota, A.; Bukowska, J. Surf. Sci. 2003, 532, 227-232.
CrossRef - Zhao, G.; Stevens, S.E. Biometals. 1998, 11, 27-32.
CrossRef - Liz-Marzan, L.M.; Lado-Tourino, I. Langmuir. 1996, 12, 3585-3589.
CrossRef - Pileni, M.P. Pure. Appl. Chem. 2000, 72, 53-65.
CrossRef - Sun, Y.P.; Atorngitjawat, P.; Meziani, M.J. Langmuir. 2001, 17, 5707-5710.
CrossRef - Henglein, A. J. Phys. Chem. B. 1993, 97, 5457-71.
CrossRef - Henglein, A. J. Chem. Mater. 1998 10, 444-446.
CrossRef - Henglein, A. Langmuir, 2001,17,2329-2333.
CrossRef - Jae, Y.S.; Beom, S.K. Bioprocess. Biosyst. Eng. 2009, 32:79-84.
CrossRef - Klaus, T.; Joerger, R.; Olsson, E.; Granqvist, C.G. J. Proc. Natl. Acad. Sci. USA. 1999, 96, 13611-13614.
CrossRef - Shiv Shankar, S.; Rai, A.; Ahmad, A.; Sastry, M. J. Colloid. Interface. Sci. 2004, 275, 496-502.
CrossRef - Gardea-Torresdey, J.L.; Parsons, J.G.; Gomez, E.; Peralta-Videa, J.; Troiani, H.E.; Santiago, P.; Yacaman, M.J. Nano. Lett. 2002, 2,397-401.
CrossRef - Chandran, S.P.; Chaudhary, M.; Pasricha, R.; Ahmad, A.; Sastry, M. Biotechnol. Prog. 2006, 22, 577-83.
CrossRef - Huang, J.; Li, Q.; Sun, D.; Lu, Y.; Su, Y.; Yang, X.; Wang, H.; Wang, Y.; Shao, W.; He, N.; Hong, J. Nanotech. 2007, 18, 105-104.
- Ankamwar, B.; Chaudhary, M.; Sastry, M. Syn. Reac. Inorg. Met. Org. Nano. Met. Chem. 2005, 35,19-26.
CrossRef - Govindan, S.; Viswanathan, S.; Vijayasekaran, V.; Alagappan, R. J. Ethnopharm. 1999, 66, 205-10.
CrossRef - Silva, C.J.S.M.; Ubitz, G.G.; Arthur, C.P. J. Chem. Technol. Biotech. 2006, 81, 8-16.
CrossRef - Adinarayana, K.; Suren, S.; Biochem. Eng. J. 2005, 27,179-190.
CrossRef - Iravani, S.; Korbekandi, H.; Mirmohammadi, S.V.; Zolfaghari, B. Res. Pharm. Sci. 2014, 9, 385–406.
- Singh, M.; Sinha, I.; Mandal, R.K. Mat. Let. 2009, 63, 425-427.
CrossRef - Jiang, X.; Chen, W.; Chen, C.; Xiong, S.; Yu, A. Nano. Res. Lett. 2011, 6, 32.
- Sun, Q.; Cai, X.; Li, J.; Zheng, M.; Chen, Z.; Yu, C.P. Col Surf A: Phys. Eng. Asp. 2014, 444, 226-231.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.









