Spectroscopic Studies of Charge Transfer Complexes of Some Amino Acids with Copper (II)
Fethi Abed1*,Houari Rayah1, Rachid Bouamrane2 and Ali Hasoon Al-Taiar2
1Laboratory of Chemistry, Gas Refining and Petrochemical Department, Arzew School, Sonatrach/Algerian Petroleum Institute (IAP), BP 172 Ain El Biya, Oran 31213, Algeria.
2Department Physics, Faculty of Science, Laboratory surface and structure of materials treatment,University of Science and Technology P.O.Box 29031 U.S.T.O. (MB) Oran 31036, Algeria.
DOI : http://dx.doi.org/10.13005/ojc/310240
Article Received on :
Article Accepted on :
Article Published : 22 Jun 2015
In this study, UV-Visible absorption studies of these complexation reactions accured between amino acids have electron donors and Copper (II) have year electron acceptor are studied. From these reactions some parameters such have, reaction constant equilibrium, (KAD), molar extinction coefficient, ( AD), absorption bands energy, (ECT), for the produced organo-metallic compounds, the ionization potential of the electron donor, (ID) and the reactions Gibbs free energy exchanges, ( G°), were calculated and discussed.
KEYWORDS:UV-Visible spectrophotometry; Organo-metallic complex; Benesi-Hildebrand equation; Molarextinction coefficient; Aminoacids; Copper (II)
Download this article as:| Copy the following to cite this article: Abed F, Rayah H, Bouamrane R, Al-Taiar A. H. Spectroscopic Studies of Charge Transfer Complexes of Some Amino Acids with Copper (II). Orient J Chem 2015;31(2). |
| Copy the following to cite this URL: Abed F, Rayah H, Bouamrane R, Al-Taiar A. H. Spectroscopic Studies of Charge Transfer Complexes of Some Amino Acids with Copper (II). Available from: http://www.orientjchem.org/?p=9381 |
Introduction
Metals are mainly toxic to living room organisms and they tighten to accumulate in their living room tissues. In recent years, there has been has growing public concern and awareness of toilets problems. This is attributed to the great potential human health risks associated with toilets pollution.
Amino acids forming essential shares of human body represent have very interesting class of compounds because of to their utility:
Normal metabolic processes involving all living room organisms and Protection of the organism from toxic effects these metals. The importance of these amino acids is due to their ability to form stable complexes with various metallic cations by chelating reactions[i]-5.
Many analytical methods use molecular absorption spectrophotometry of co-ordination compounds for the determination of metal ions in solution. This milked study by electronic spectroscopy of the behavior of the aliphatic amino acids with respect to Copper (II) in aqueous solution at 298 K.
The spectroscopic parameter of the association constant,KAD, Gibbs energy change,
ΔG°, were calculated, as well as molar extinction coefficients, AD, by the use of the equation of Benesi-Hildebrand.1. Moreover the energy of absorption (ECT), as well as the potentials of ionization of the ligands(ID),will be established in order to better circumscribe the behavior of charge transfer complex of five amino acids with Copper (II)6-12.
Experimental
General
Copper(II) Nitrate [Cu (No3)2,5H2O] from Merck, amino acids from Fluka (purity 99 %).All the solution was prepared in 100 cm3 in double distilled water.
The UV-visible spectra were measured using Perkin – Elmer lambda 20 and UV-visible Shimadzu 1202 (Shimadzu Europa Gmbh) and a quartz cuvette (1-cm path length). The weightings are carried out in a Sartorius balance CPA124 S.
General procedure
Electronic spectra of various compounds were recorded between 200 and 1100 nm, in the aqueous solution of variable concentrations of donor and acceptor 10-2 and 10-4 mole.l-1 and maximum wavelength λ max were characterizing at 298 K.The association constant KAD, as well as molar extinction coefficients εAD, of each complex were calculated using the equation of Benesi-Hildebrand.113.
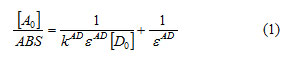
Where:
ABS: absorbance of the charge transfer band,[A0]: initial concentration of the electron acceptor, [D0]: initial concentration of the electron donor,KAD: association constant of charge transfer complex in solution, εAD: molar extinction coefficient of charge transfer complex.
The ionization potential of the electron donors were calculated from the position of charge transfer (CT) bands for iodine complexes using the following equations: 10-17
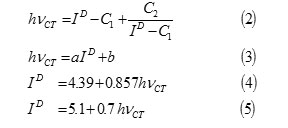
hvCT: Absorption band energy of charge transfer complex, and h : Plancks constant,
C1 =5.2 eV,
C2 =1.5 eV2,
a = 0.67 or 0.87,
b=-1.9 or -3.6.
Where hvCT is the absorption band energy of the CT complex in e.v., ID is ionization potential of the electron donor in ev., C1, C2, a and b in Equations.2 and 3 are constants and their values for the iodine acceptor in CCl4are as follows:
Results and Discussion
All the acids amines have the property of absorptive the light ultraviolet. The absorption band of the Copper (II) with various amino acid donors and their charge complex were measured in aqueous solution and are tabulated in Table.1.All complexes exhibit new absorption bands in the visible region their wave lengths are 740,750,750, 750 and 760 nm for the D1 to D2 complexes, respectively, wile 810 nm band of the acceptor disappeared.
 |
Table1: Wavelengthλmax(nm)of the Copper complexes Cu (II)with various amino acid donors, in aqueous solution. Click here to View table |
The values of association constant KAD and the coefficients of molar extinction AD are deduced by plotting:
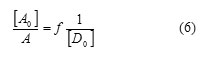
By the use of the equation of Benesi–Hildebrand.113.The values of KAD, ε AD and Gibbs energy changes ΔG° for all complexes, were obtained as shown in Fig.1-5 and are reported in Table.2.
Table2: Values of association constants, molar extinction coefficient, and Gibbs energy change for each complex of amino acids with Cu (II) in aqueous solution.
|
Donors |
KAD(l.mole-1) |
AD(l.cm-1.mole-1) |
– G°(KJ. mole-1) |
|
Glycine (D1) |
1241 |
19.402 |
17,847 |
|
DL – Alanine (D2) |
1384 |
17.683 |
18,119 |
|
Leucine (D3) |
1261 |
19.936 |
17,885 |
|
Isoleucine (D4) |
1354 |
20.429 |
18,064 |
|
Valine (D5) |
1559 |
20.653 |
18,417 |
![Fig.1: Plot of ([A]0/ABS) versus(1/[D]01) for the complex [Cu (II)/Glycine],](http://www.orientjchem.org/wp-content/uploads/2015/06/Vol31_No2_Spe_FET_Fig1-150x150.jpg) |
Figure1: Plot of ([A]0/ABS) versus(1/[D]01) for the complex [Cu (II)/Glycine], |
![Fig.2: Variation Plot of([A]0/ABS) versus (1/[D]02) for the complex [Cu (II)/DL Alanine],](http://www.orientjchem.org/wp-content/uploads/2015/06/Vol31_No2_Spe_FET_fig2-150x150.jpg) |
Figure2: Variation Plot of([A]0/ABS) versus (1/[D]02) for the complex [Cu (II)/DL Alanine], Click here to View figure |
![Fig.3:Plot of ([A]0/ABS) versus (1/[D]03) for the complex [Cu (II) /Leucine]](http://www.orientjchem.org/wp-content/uploads/2015/06/Vol31_No2_Spe_FET_Fig3-150x150.jpg) |
Figure3: Plot of ([A]0/ABS) versus (1/[D]03) for the complex [Cu (II) /Leucine] Click here to View figure |
![Fig.4: Variation Plot of([A]0/ABS) versus (1/[D]04) for the complex [Cu (II) /Isoleucine].](http://www.orientjchem.org/wp-content/uploads/2015/06/Vol31_No2_Spe_FET_Fig4-150x150.jpg) |
Figure4: Variation Plot of([A]0/ABS) versus (1/[D]04) for the complex [Cu (II) /Isoleucine]. Click here to View figure |
![Fig.5:Plot of([A]0/ABS) versus (1/[D]05) for the complex [Cu (II)/Valine].](http://www.orientjchem.org/wp-content/uploads/2015/06/Vol31_No2_Spe_FET_Fig5-150x150.jpg) |
Figure5: Plot of([A]0/ABS) versus (1/[D]05) for the complex [Cu (II)/Valine]. Click here to View figure |
The study of the compared values of the various constants of balance, KAD for the complexes formed starting from the aliphatic amino acids, reflects a greater stability for that formed with D7; the complex [ D1 / Cu(II) ] proves to be the least stable.
Molar absorptivities, ε AD, obtained from Equation.1, are 19.402, 17.683, 19.936, 20.429 and 20.653. I.cm–1mol–1for complexes D1 to D5, as shown in Table.2, wile the values of KAD, are given in Table.2. These 1241, 1384, 1261, 1354 and 1559 (dm3.mole-1), which make it possible to establish the order of stability for the same complexes as following:
[D5/Cu (II)] > [D2/Cu (II)]> [D4/Cu (II)]> [D3 /Cu (II)]>[D1/ Cu (II)]
The comparison of the stability of the complexes [D4/Cu (II)] and [D3/Cu (II)] shows a stability appreciably more significant for the complex [D4 /Cu (II)] because of the grouping methyl substituted for the carbon β which is closer compared to Cα carbon.
The stability compared between the complexes [D1/Cu (II)] and [D2/Cu (II)] is clearly in favor of the complex [D2/Cu (II)]. This is due to the presence of the grouping methyl (inductive donor) substituted for carbon C α.
Moreover, the Gibbs energy changes for the all complex formation reactions calculated from the values of KAD, are -17,847, -18,119, -17,885, -18,064 and -18,417 KJ. mole-1,as shown in Table.3. This shows that the spontaneity of these reactions follows the trend:
[D1/Cu (II)]> [D3/Cu (II)] > [D4/Cu (II)] > [D2 /Cu (II)] > [D5/ Cu (II)]
The absorption band energies are calculated by the equation: ECT =hvmax. (6), the results for each complex are deferred in Table.3.
The values of ionization Potential ID of the various amino acids,complexing Copper (II) in the aqueous solution calculated by applying equations.4 to 7 whenethe average values of ID range between 7,686 and 7,897 eV, are reported in Table.4.18-21
This study opens new prospects when with the use of these various complexes like catalysts in other reactions of organic chemistry. New studies are undertaken in this direction which aims at correlating the catalytic results obtained with the structure of the organometallic complex. The whole of the results obtained enables us to consider new possibilities in which the amino acids could, through their metal complexes, to constitute good agents extractants in the extractions liquid liquid.
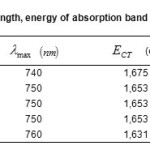 |
Table3: Values of wavelength, energy of absorption band of the complexes ECT(eV), |
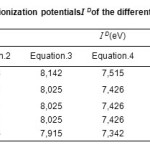 |
Table4: Calculated values of the ionization potentials I D of the different amino acid, in aqueous solution. Click here to View table |
Conclusion
In this work, the study carried out relates to the behavior complexing of the various aliphatic amino acids with Copper (II) in the aqueous solution with 25 °C, by the means of the electronic spectroscopy.
This study made it possible to release a great number of physical parameters characterizing these complexes:
The spectroscopic results us suggest that the whole of
the complexes are of stoichiometry Ml.
The use of the equation of Benesi-Hildebrand made it possible to establish for the various formed complexes their constants of formation KAD, molar extinction coefficient εAD, as well as the variations of their Gibbs energy change Δ G°.
The study of the compared values of the various association constants,KAD for the complexes reflects the greatest stability for that formed with D5, followed that with the valine. The complex [D1/Cu (II)] finally proves to be the least stable.
References
- R.Atkins, G. Brewer,E.Kokot and G.M. Mockler, Inorg. Chem., 1985. 24, 127–129
- R. Yuan, Y. Chai, D. Liu, D. Gao, J. Li and R. Yu, Anal. Chem., 1993.65,2572–2575
- R. Ramesh, P.K. Suganthy and K. Natarajan, Synth. React. Inorg.Met.-Org. Chem. 1996.,26, 47–60
- R.A. Siddiqui, P. Raj, A.K.A. Saxena and S.K. Dixit, Synth. React. Inorg.Met.-Org. Chem., 1996.26, 1189–1203
- S.Y. Ohashi, Bull. Chem. Soc. Japan;,1997.,70, 1319–1324
- A.H. Al-Taiar, S.K. Shubber and A. Megherbi, Turk. J. Chem. 2002.,26,351–355
- A.H. Al-Taiar, M.A. Al-Ekabe, F.A. Al-Mashadaneand M. Hadjel,J. Coord. Chem. 2002.,55, 1143–11468.
- S.H. Hastings, J.L. Franklin, C. Schiller and F.A. Matsen, J. Am. Chem.Soc. 1953.,75, 2900–2905
- H.M. McConnell, J.S. Ham and J.R. Platt, J. Chem. Phys., 1953.21,66–70
- R.S. Mulliken and W.B. Person, Ann. Rev.Phys. Chem., 1962.13, 107–126
- F.A. Matsen, J. Chem. Phys. 1956.,24, 602–606
- H. Kuroda, Nature, 1964.201, 1214–1215
- H.A. Benesi and J.H. Hildebrand,J. Am. Chem. Soc. 1949.,71, 2703–2707
- R. Foster and I. Horman, J. Chem. Soc. 1966. (B), 1049–1053
- F.M. Menger and M.L. Bender, J. Am. Chem. Soc., 1966.,88, 131–137
- K.J. Laidler, Chemical Kinetics, 2nd edn., McGraw-Hill, London 1965.,. 19–21
- P. Morlaës and J-C. Morlaës, Cinétique Chimique, Vuibert, St-Germain, France, 1949).. 166–168
- K.R. Bhaskar, R. K. Gosavi and C.N.R. Rao, Trans. Faraday Soc. 1966.,62, 29–38
- K.R. Bhaskar, S.N. Bhat, A.S.N. Murthy and C.N.R. Rao, Trans. Faraday Soc. 1966.,62, 788–794
- H. Yada, J. Tanaka and S. Nagakura, Bull. Chem. Soc. Japan 1960.,33, 1660–1667
- R.S. Mulliken, J. Chim. Phys. 1964.,61, 20–38

This work is licensed under a Creative Commons Attribution 4.0 International License.









