Highlights of the Meeting on “Horizontal Gene Transfer and the Last Universal Common Ancestor” Held at the Open University, Milton Keynes, UK
Sohan Jheeta
Network of Researchers on Horizontal Gene Transfer and the Last Universal Common Ancestor, 1 Scott Hall Crescent, Leeds, LS7 3RB, U.K.
DOI : http://dx.doi.org/10.13005/ojc/290459
Article Received on :
Article Accepted on :
Article Published : 11 Jan 2014
This paper summarises the oral presentations, their outcomes and conclusions drawn from the meeting entitled: ‘Horizontal Gene transfer and the Last Universal Common Ancestor’ which was held at the Open University, Milton Keynes, UK, on the 5th and the 6th September 2013.
KEYWORDS:Horizontal gene transfer;HGT;Lateral gene transfer;LGT;Phylogenetic tree of life;Transformation; Gene transfer agent (GTA); Last universal common ancestor (LUCA)
Download this article as:| Copy the following to cite this article: Jheeta S. Highlights of the Meeting on “Horizontal Gene Transfer and the Last Universal Common Ancestor” Held at the Open University, Milton Keynes, UK. Orient J Chem 2013;29(4) |
| Copy the following to cite this URL: Jheeta S. Highlights of the Meeting on “Horizontal Gene Transfer and the Last Universal Common Ancestor” Held at the Open University, Milton Keynes, UK. Orient J Chem 2013;29(4). Available from: http://www.orientjchem.org/?p=1557 |
INTRODUCTION
One of the first of its kind, this meeting, held on the 5th and the 6th September, was designed to investigate and evaluate the importance of ‘horizontal gene transfer’ (HGT) during the period of the ‘last universal common ancestor’ (LUCA) prior to the emergence of the three domains (Archaea, Bacteria and Eukarya) of life and their becoming separate branches of the phylogenetic tree (Figure 1). The idea of the tree of life is not new as it was first suggested by Charles Darwin who reasoned that every living species was a product of a species that came before it and that they all emanated from a single common organism (Figure 2). The meeting brought together an international gathering of scientists from Japan, India, Israel, Russia, USA, Canada, Germany, Denmark, Italy, Spain as well as participants from the UK (Figure 3). With such an assortment of eminent researchers under one roof, the meeting was both energetic and engaging and generated a lively debate and exchange of ideas. The synopses of the presentations are outlined in this report.
Prof. J. Thomas Beatty (University of British Columbia) presented: ‘Gene transfer agents in prokaryotes: viral progenitors, or revamped phages?’
Prof. Beatty and his group study the induction of the prophage-like gene transfer agent RcGTA, of the purple non-sulfur bacterium Rhodobacter capsulatus. Several pathways that affect the production of extracellular RcGTA particles have been discovered. One pathway involves the response-regulator CtrA, which is essential for transcription of RcGTA structural genes; a second pathway transmits a quorum-sensing signal, produced by a homoserine lactone synthase, which is needed for maximal induction of transcription of RcGTA genes; and a third pathway requires the sensor-kinase homologue CckA for maturation of RcGTA particles and release from cells. It was found that RcGTA particles are released by cell lysis and that a small percentage of wild type cells induce RcGTA gene expression and go on to essentially commit suicide. Induction of RcGTA genes appears to be controlled by a stochastic process involving changes in gene transcription, as opposed to a genetic modification.
Genome sequences show that most prokaryotes contain prophage-like gene clusters, but possible relationships to gene transfer agents (GTAs) were unclear. Beatty and coworkers established that RcGTA-like gene clusters are widespread but found exclusively within the alpha-proteobacteria. A phylogenetic tree comparing 16S rRNA and RcGTA gene homologue sequences indicates that these two types of gene have evolved in concert. It appears that a single RcGTA ancestor arose in the alpha-proteobacteria after divergence of this line from other prokaryotes. The similarity of 16S rRNA and RcGTA-like protein sequence trees in of present-day species indicates that the transmission of RcGTA-like genes was largely vertical, as for most cellular genes, with little or no HGT. They suggested that GTAs have arisen independently in multiple prokaryotic lines, and that at least RcGTA arose long after the initial divergence of prokaryotes into ancestors of present-day species.
Prof. Matxalen Llosa, University of Cantabria, Spain presented: ‘Bacterial conjugation: a protein secretion system adapted to DNA transfer, or vice versa?’
Prof. Llosa discussed the molecular mechanism of bacterial conjugation, whereby DNA segments are transferred horizontally from one extant bacterial cell to another living bacterial cell via the multiprotein complex: T4SS, a type IV secretion system normally involved in protein secretion and it is derived from DNA transfer machineries. The same channel [T4SS] is used to transport a DNA strand from one cell to another and it occurs when DNA replicase excises a DNA strand; the strand and replicase bind to form a nucleoprotein complex; it is this complex which is transferred to another extant cell via T4SS.
She proposed a coupling protein (CP) that both mediates substrate recognition and DNA transfer through the T4SS. Additionally, bacterial conjugation comes about by merging a rolling-circle replication system with a T4SS. It was further proposed that CP may be a key factor in the evolution of these secretion systems, as it interacts with both the secreted substrate and the T4SS with different specificities.
Prof. Armen Mulkidjanian (University of Osnabrueck, Germany/ Lomonosov University, Moscow, Russia) presented: ‘Energetics of the first cells and the Last Universal Cellular Ancestor (LUCA)’
The broad premise of Prof. Mulkidjanian’s presentation was the importance of K+, Mg2+, Zn2+, Mn2+ and phosphate ions in the origin of life on Earth. This is because these ions, in modern biology, are essential for the ‘proper’ function of a large number of ubiquitous proteins and functional systems such as ribosomes, diverse ATPases and GTPases, rotary membrane ATP synthases, which depend on high levels of these ions; from this premise it can be surmised that the same ions with similar concentrations to those of modern biology were needed for supporting the LUCA. The environmental setting for the origin of life, where these ions could have been present, was thought to have been the vapour-dominated zones of inland geothermal systems rather than the underwater hydrothermal vents on the sea floor. Prof. Mulkidjanian also put forward that any LUCAs were likely to be consortia of replicating life forms, not necessarily with the same genomes as each other, rather than a single LUCA organism. HGT and the sharing of gene products and metabolites would have been vital for the survival of such primordial consortia which eventually gave rise to the 3 domains of life.
Prof. John Allen (Queen Mary, University of London) presented: ‘Coupling between information processing and redox chemistry. Cellular co-location of energy transduction with genome function.’
Prof. Allen proposed two hypotheses. The first is ‘Co-location for Redox Regulation of gene expression’ (CoRR) and its relation with bacterial redox regulatory systems. CoRR states that a biological energy generating apparatus is under the control of the genomic informational system and vice versa - such coupling channels are to be found in both mitochondria and chloroplasts, which are derived from the LUCA. The second hypothesis, the ‘redox switch hypothesis’ predicts that the first oxygen-evolving organism, a cyanbacterium, was the descendant of an autotrophic bacterium that performed anoxygenic photosynthesis using hydrogen sulphide. In this proposed ancestor, the revolutionary advent of free molecular oxygen on Earth resulted from a mutation that caused a failure of redox regulation, leading to expression of genes for both photosystem I and photosystem II reaction centres.
Dr. Søren Overballe-Petersen (Centre for GeoGenetics, Natural History Museum of Denmark) presented: ‘Natural transformation of bacteria by highly fragmented, damaged and ancient DNA.’
DNA from both extant and dead cells can survive for thousands of years, forming a natural pool of fragmented extracellular DNA. Dr. Overballe-Petersen showed that this pool of DNA, although fragmented into various lengths (e”20 bp), retains the capacity for natural transformation – for example he demonstrated that 40,000 years old ‘short’ DNA can naturally transform bacteria so that cells can either acquire functional genetic signatures from the distant past or it could produce completely new variants of already functional genotypes. Such results provided a mechanistic support to the hypothesis that HGT probably played an important role during the evolution of LUCA en route to the formation of the three domains that are observed in phylogenetic studies today.
Prof. Vinod Gupta (C.M.D. Post Graduate College, India) presented: ‘Photosynergistic collaboration of non-linear process at mesoscopic level and emergence of a minimal protocell-like photoautotrophic supramolecular assembly “Jeewanu” in a simulated possible prebiotic atmosphere.’
Prof. Gupta showed the result of his experiment which formed proto-cell like entities, referred to as Jeewanu, which could be made from a sterile mixture of various chemicals (ammonium molybdate, diammonium phosphate, formaldehyde and ‘biological minerals’), when exposed to sunlight, the reaction produced vesicles surrounded by semi-permeable membranes. These Jeewanu were bluish in colour, double layered spheres which were able to multiply by budding and they were able to carry out photolysis of water – a process akin to photosynthesis; such experimentation has yet to be verified independently. If anyone is interested in validating these findings, please contact Prof. Gupta as he would be only too pleased to provide details of the experiment upon request via: vkgcmd@gmail.com.
Prof. Addy Pross (Ben Gurion University of the Negev, Israel) presented: ‘Seeking the evolutionary roots of horizontal gene transfer.’
Prof. Pross explained the roots of HGT from the point of view of ‘systems chemistry’- a relatively new area of science. Complex self-assembling systems underpin all life and the new ‘systems chemistry’ is the modern explanatory tool. This area of chemistry deals with the emergent properties of self-replicating molecular systems and the chemical networks they establish. In order to explain this Prof. Pross introduced the concept of dynamic kinetic stability (DKS), a stability kind which is associated specifically with replicating systems, unlike the more widely applicable thermodynamic stability, normally associated with ‘regular’ chemistry. DKS is likely to have begun with small replicating networks, possibly involving RNA molecules, building slowly, giving rise to increasingly complex systems and ultimately culminating in the formation of living entities. Moreover, by examining and predicating the emerging properties of these replicating molecules/networks, some insight can be gleaned into the earliest phases of the evolutionary process. Thus HGT would likely have been rampant, LUCA presumed to have been communal rather than individual, and what is termed the Darwinian threshold is attributed to the beginnings of speciation, when the 3 domains of life began to diverge.
Prof. Prakash Joshi (Rensselaer Polytechnic Institute) presented: ‘Prebiotic RNA synthesis by montmorillonite catalysis: significance of mineral salts.’
Prof. Joshi described the experimental basis for the formation of oligomers of RNA molecules on the surface of montmorillonite clay minerals in the presence of different anions and cations. His experiment can be seen as groundbreaking in that such oligomers form the basis of the ‘RNA world’ hypothesis; a pathway in which the first living entities (biopolymers) were thought to be RNA, which predated the DNA/protein entities. This is because RNA molecules have dual properties whereby they can self-catalyse reactions as well as acting as repositories of inheritable biological information. It has been shown by other researchers that even a pentaribonucleotide (GUGGC) is able to carry out very important functions including aminoacylation, transacylation and peptide synthesis.
Prof. Michael Syvanen (University of California at Davis) presented: ‘Why LUCA is no longer needed as part of evolutionary theory?’
Prof. Syvanen made a compelling argument that HGT obviates the need for a postulated LUCA. This hypothesis is based on the proposition that widespread HGT occurred during the period of the emergence of the very earliest forms of life and has occurred at a frequency sufficient to account for those genes found in life’s three domains. The evidence presented was in the form of highly conserved genes, which appear to be younger than the taxonomic groups in which they are found today. These genes include those that are involved in the biosynthesis of arginine and tryptophan as well as their respective tRNA ligases. The suggestion here is that the modern genetic code was not established until well after the major domains had already differentiated. Thus the common ancestry of these two amino acids can be explained by stipulating that there were unknown numbers of evolving ‘collective LUCAs’ from which it could have been acquired via HGT rather than from the stipulated single entity called LUCA; that is, traditionally, LUCA represented a single root to the tree of life and therefore represented a bottleneck. Therefore, the concept of LUCA, as a single entity and predecessor of all living entities on Earth today, does not explain the universal distribution of the above mentioned amino acids. In essence, Prof. Syvanen maintains that a much more plausible scenario would be the ‘collective LUCA.’
Dr. Duccio Medini (Novartis Vaccines Research Centre, Siena) presented: ‘Length-modulation of horizontal gene transfer explains contradictory population structure in pathogenic bacteria
In essence Dr. Medini’s research is very important from the future point of view of bacterial population (colonal, panmictic and intermediate structures) involving HGT and its role since the LUCA era to date in shaping bacterial population structures. He found in a model bacterial species, Neisseria meningitis, gene-conversion events to be longer within population compartments (phylogenetic clades) rather than between compartments, suggesting a DNA cleavage mechanism associated with the species phylogeny. His group also identified 22 restriction modification systems, probably acquired by HGT from outside of the species/genus, whose distribution in the different strains coincides with the phylogenetic clade structure. They showed with in-silico evolution experiments that the length-modulation of DNA exchange provided by restriction modification systems is sufficient to generate the observed population structure. These findings have general implications for the emergence of lineage structure and virulence in recombining bacterial populations, and could provide an evolutionary framework for the population biology of other species (and possibly LUCA) showing contradictory population structure and dynamics.
Prof. Nigel Goldenfeld (University of Illinois at Urbana-Champaign) presented: ‘How a “collective LUCA” drove the rapid evolution of early life: clues from the canonical genetic code.’
Prof. Goldenfeld’s main premise of the presentation was that the genetic code is not a random state of affairs, but, instead, is ‘hard wired’ into the evolution of life even before the advent of LUCA and that it is both universal and also nearly optimally ‘tweaked’ such that it has certain error minimisation properties. More importantly, the relics of early life are still present in structure of the modern day canonical genetic code. The hard wiring arose as a result of high levels of HGT between individual entities within the ‘proto-LUCA’ community, during the time of life’s early evolution; this horizontal gene transfer process of core cellular machinery dominated vertical evolution, with the eventual formation of the three distinct domains of life. Thus, the central theme of the presentation was that life arose in communal settings as opposed to emerging via a single entity route. Prof. Goldenfeld used digital simulations to show that principally, the genetic code’s universality and optimality occurred rapidly due to HGT and Lamarckian evolution, within the collective LUCA and that the code had the ‘cunning’ property of being ‘tamper-proof.’
Dr. Sohan Jheeta (Independent Researcher and Chairman: NoR HGT & LUCA) presented: ‘Hypothetical consideration: tRNAs are the kings of the hill.’
The discovery that pentaribonucleotide (GUGGC) is able to carry out aminoacylation, transacylation and peptide synthesis (therefore it acts as a catalyst) raises the profile of RNA in relation to the major part it played in the origin of life. RNA molecules were present just prior to the emergence of the ‘hypothetical’ cell now commonly referred to as the ‘last universal common/cellular ancestor’ (LUCA). The LUCA is thought to be the predecessor of the three domains of life as has been shown by the phylogenetic tree of life. Dr. Jheeta put forward a hypothesis that suggested that, of the three prominent RNA molecules (mRNA, rRNA and tRNA) used by life on Earth today, tRNA may have been the molecule which acted as the carrier of genetic information; was responsible for catalysing the early reactions; and carried anticodons at one end as well as activated amino acids at its other end (ie 32 ). tRNA was involved in the formation of the earliest types of ribonucleoproteins and formed early ‘rudimentary’ ribosomes which gave rise to present day ribosomes.
Panel open Discussion
This session was chaired by Dr.Jheeta and the participants were: Profs Armen Mulkidjanian, Addy Pross, Michael Syvanen and Nigel Goldenfeld and the question they discussed was: ‘What part did HGT play during the evolution of LUCA leading to the three domains?’
During the conference it quickly became clear that, perhaps, LUCA ought to be more clearly defined because according to the phylogenetic tree of life, there was a point where all domains of life went through a bottleneck created by a single living entity: LUCA. This is not an inconsequential issue as many scientists put a lot of credence on the tree and believe that life had a singularity of origin. However, this singularity was seen as a moot point since the panel agreed, as result of their research, that there is a high degree of probably that the LUCA may not have been one single entity, but rather consortia of LUCAs (as Prof. Mulkidjanian declared) as summarised in the conclusion section.
Where do we go from here?
The final session of the day, which was also chaired by Dr. Jheeta, focussed on the question: ‘where do we go from here?’ This issue was debated thoroughly by both the panel and the other participants. There were ultimately two main outcomes: that there is a need to hold annual meetings to discuss, consider and draw conclusions in relation to HGT and the LUCA; and also that there is a particular requirement to form a group in order to co-ordinate this annual activity. It was decided that the group be called the ‘Network of Researchers on Horizontal Gene Transfer and the Last Universal Common Ancestor’ or NoR HGT & LUCA (working website: www.hgts.co.uk).
Conclusion
The findings concerning HGT, the LUCA and the phylogenetic tree of life meeting were as follows: HGT was rife and occurring on a gigantic scale, reorganising the LUCA’s genetics, which then ultimately led on to the emergence of life. The LUCA was a highly sophisticated entity to the point that it was construed to be alive and was RNA based, implying that it was a single entity as supposed by ardent biologists; however, the meeting concluded that it was highly possible that a community of diverse LUCAs co-existed in the vicinity of one another. To begin with LUCAs were less sophisticated and could be referred to as proto-LUCAs which exchanged genetics via HGT. Also it was generally agreed that the word ‘common’ in the last universal common ancestor should be changed to the word ‘cellular’ as the ‘common’ does not explain the origin of viruses, since both LUCA and viruses had a parallel origin, noting that viruses also had a ‘hand’ in the emergence of life. As with the LUCA, the phylogenetic tree of life is also a theoretical construct to explain the origin of life, but it was demonstrated by Prof. Michael Syvanen that, by reworking the phylogenetic tree, it may not be an appropriate device when describing the origin of its three branches.
Another conclusion of the meeting was that it was agreed that HGT is also the essence of Lamarckian evolution rather than Darwinian and so it was via such interactions that the genetic code in the very early LUCA was reorganised, leading eventually to the seemingly rapid formation of the 3 domains of life.
Acknowledgements
I would like to thank all the contributors for submitting such an array of interesting and stimulating abstracts. In addition, I would like to thank my co-conveners, Dr. Martin Dominik (University of St. Andrews), Prof. Nigel J. Mason as well as the Open University for hosting the meeting entitled: ‘Horizontal Gene Transfer and the Last Universal Common Ancestor’ on 5–6 September 2013, from which some of the material in this publication is drawn.
Conflict of Interest
The authors declare no conflict of interest.
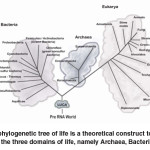 |
Fig. 1: The phylogenetic tree of life is a theoretical construct to explain the emergence of the three domains of life, namely Archaea, Bacteria and Eukarya Click here to View figure |
 |
Fig. 2: Transmutation of Species as depicted on page 36 of Darwin’s notebook B Click here to View figure |
 |
Fig. 3: scientists who attended the first Horizontal Gene Transfer and the Last Universal Common Ancestor conference at the Open University, Milton Keynes, UK, on 5-6 September 2013 Click here to View figure |

This work is licensed under a Creative Commons Attribution 4.0 International License.










