Antioxidant, Antibacterial and Antifungal Properties of Black Pepper Essential Oil (Piper nigrum Linn) and Molecular Docking and Pharmacokinetic Studies of its’ Major Component.
Rajia Sultana1* , Md. Din Islam1
, Md. Din Islam1 , Fazria Tanjum2
, Fazria Tanjum2 , Mohammad Mostafizur Rahman2
, Mohammad Mostafizur Rahman2 , Md. Aminul Haque2
, Md. Aminul Haque2 , Rashadul Hossain1
, Rashadul Hossain1
1Department of Chemistry, Chittagong University of Engineering and Technology, Chittagong 4349, Bangladesh.
2Department of Chemistry, Jagannath University, Dhaka 1100, Bangladesh.
Corresponding Author E-mail: rajiasultana@cuet.ac.bd
DOI : http://dx.doi.org/10.13005/ojc/380630
Article Received on : 07 Nov 2022
Article Accepted on : 17 Dec 2022
Article Published : 28 Dec 2022
Reviewed by: Dr. Sovan pattanaik
Second Review by: Dr. Ioana Stanciu
Final Approval by: Dr.Vandana Magarde
The present study aimed to investigate chemical composition of essential oil (EO) from black pepper extract through steam distillation and evaluate by GCMS with in vitro antimicrobial and antioxidant activities and in silico studies. In total, thirteen volatile compounds identified by GCMS analysis. Among them, main components are d-norandrostane (14.874%), Valencene (13.297%), 1H-3a-7-Methanoazulene octahydro-1,9,9-trimethyl-4-methylene- (11.591%), (-)-spathulenol (8.193%), aromandendrene (8.398%), and naphthalenedecahydro-4a-methyl-1-methylene-7-(1-methylethylidene) (7.794%). The EO extracted from black pepper, displayed moderate antibacterial activity against ten bacterial strains (two and eight numbers of Gram-positive and Gram-negative, respectively) compared with Ceftriaxone as standard. In case of antifungal study, the EO exhibited a greater zone of inhibition with 13.7±1.5 mm against Trichodermal harzianum, compare to Amphotericin B as standard (11.7±1.5 mm). The results of antioxidant efficacy of extracted EO revealed good activity with IC50 value 35.83±2.92 μg/mL as compared to standard ascorbic acid (27.34±1.86 μg/mL). In silico studies satisfy the experimental values.
KEYWORDS:Antimicrobial activities; Antioxidant activities; Black pepper; Essential oil; GCMS; Molecular docking
Download this article as:| Copy the following to cite this article: Sultana R, Islam M. D, Tanjum F, Rahman M. M, Haque M. A, Hossain R. Antioxidant, Antibacterial and Antifungal Properties of Black Pepper Essential Oil (Piper nigrum Linn) and Molecular Docking and Pharmacokinetic Studies of its’ Major Component. Orient J Chem 2022;38(6). |
| Copy the following to cite this URL: Sultana R, Islam M. D, Tanjum F, Rahman M. M, Haque M. A, Hossain R. Antioxidant, Antibacterial and Antifungal Properties of Black Pepper Essential Oil (Piper nigrum Linn) and Molecular Docking and Pharmacokinetic Studies of its’ Major Component. Orient J Chem 2022;38(6). Available from: https://bit.ly/3WLJzYP |
Introduction
Many plant extracts have been use to treat infectious diseases of human for centuries due to their therapeutic properties. Generally, most of the plants bear important chemical compounds called phytochemicals, which can generate specific physiological action against microorganisms on human body1. The treatment of infectious diseases caused by various pathogens has become an alarming threat due to the extensive use of antibiotics and the rapid development of multidrug resistance microorganisms2. Essential oil and other extracts of plants draw a greater interest due to their medicinal significance3. Essential oil extracted from different plant showed antiviral, antibacterial, antifungal, antioxidant and insecticidal properties4-5. Some essential oils used in the treatment of cancer, aromatherapy, food preservation, and fragrance industries6. Therefore, it has great importance to extract essential oils from different parts of plant i.e. fruits, leaves, roots, flowers, seeds, and barks7. Steam distillation, hydro distillation, and fermentation techniques usually employed to extract these oils from various plants8. These oils are complex mixtures of many volatile components, which are chemically nonpolar in nature. Black pepper (Piper nigrum Linn.) is a popular spice known as ‘King of Spices’ or ‘Black Gold’, and in Bangladesh known as ‘Golmorich’. It belongs to the Piperaceae family9,10 and is native to Bangladesh, India, Thailand, Brazil, China, Indonesia, Malaysia, Mexico, and Vietnam11. This widely used due to its aroma properties with nutrition and medicinal values10. In Ayurvedic, black pepper is use in the treatment of rheumatism, diarrhea, headache, dysentery, cholera, to remove excessive gas from gastrointestinal tract, and to increase the flow rate of urine. It also use for treatment of digestive problems, stomach disorders, neuralgia, and scabies12,13. Considering the above-mentioned biological importance of EOs, the present study aimed to extract essential oil from black pepper and evaluate its in vitro anti-microbiological and antioxidant activities using disc diffusion test and 1,1-diphenyl-2-picrylhydrazyl (DPPH) radical scavenging activity, respectively. In this study, 2, 8 and 5 number of Gram-positive bacteria, Gram-negative bacteria and fungal strains, respectively, were use for antimicrobial assay and in silico molecular docking and pharmacokinetic analysis performed in support of experimental results. The results present in this paper will useful to identify the biologically potent volatile compounds.
Experimental
Material and GCMS analysis
The black pepper collected from the local market of Chattogram, Bangladesh, wash with distilled water and dry under subdue sunlight for several days. The dry samples transform to powder using blender machine. The powder sample (370.0 g) subjected to extraction with n-hexane using Clevenger apparatus (Germany) for 72 hours. n-Hexane extract dried by using anhydrous Na2SO4 and followed by filter and solvent removed by using vacuum evaporator to obtain black pepper essential oil. The EO store in an airtight container and preserve in refrigerator at 8°C earlier use. Separation and analysis of essential oil perform by GCMS (gas chromatography mass spectrometer). Simadzu GC-17A gas chromatograph fitted with RTS-5MS capillary column (30cm×0.25mm diameter) coupled with a mass spectrometer (MS 2010 plus). Column temperature maintained at 260°C and packed by diethylene glycol succinate (10%). Ultra-high pure helium gas (99.99%) use as carrier with flow rate 1.0 mL/min maintaining a constant pressure (90 kPa).
Antimicrobial activity assay
The ‘disc diffusion test’ employed to determination of in vitro antimicrobial activity and Mueller Hinton agar and potato dextrose agar were use as base media14. These incubated 24 hour and monitored continually to check any contamination. The standardized test organism inoculation on incubated media by using sterile cotton bar and the filter paper discs (6 mm dia.) contain test sample at desire concentration located softly on pre-inoculated agar media surface and aerobic incubation done for 24 hour at 37°C and 48 hour at 26°C for bacterial and fungal pathogen, respectively. Each paper disc contained 25 µL of sample in DMSO that contained 300 µg of extracted EO, here dimethyl sulfoxide (DMSO) use as control. In addition, 10 µL of Ceftriaxone or Amphotericin B in DMSO was use on paper disc as standard. The Petri dishes incubated for 24 hr and the diameters of inhibition zones (mm) measured by a measuring scale. All the tests done repeated for three times. In this study, two numbers of Gram-positive bacteria named Staphylococcus aureus and Bacillus magneterium; moreover, eight numbers of Gram-negative bacteria, named Salminella paratyphi, Enterotoxigenic Escherica coli, Salminella typhi, Shigella flexneri, Shigella sonnei, Shigella boydii, Escherica coli, Shigella dysentery were used to determine antimicrobial activities. Beside this, six numbers of fungal strains namely Aspergillus niger, Panysalium notatum, Candida albican, Aspergillus flavus, Neurospora crassa, Trichodermal harzianum were used to check antifungal activity.
Antioxidant activity assay
DPPH radical scavenging method employed to find out antioxidant efficacy of EO according to a method described by Shah and co-workers15. In this method, solutions were prepared separately in ethanol with 10 mL of DPPH radical (0.1 mM), essential oil at desire concentration (31.25, 62.5, 125, 250, 500 μg/mL) and ascorbic acid as standard. 4 mL DPPH radical solution with 100 µL sample solutions were vortex and incubated at 26°C for 15 min and maximum absorbance observed at 517 nm with a blank sample (2 mL DPPH with 2 mL ethanol) by an UV spectrophotometer. The test performed three times. Inhibition (%) by the radicals calculated with equation:
Here, Abcontrol is absorbance of DPPH radical and Absample is absorbance of DPPH with essential oil. The concentration-inhibition curves utilized to determine IC50 of essential oil and ascorbic acid.
Molecular docking study
Three-dimensional crystal structure of target protein 1JIJ16, 1KZN17 and 5JBO18 of S. aureus, E. coli and T. harzianum, respectively, retrieved in PDB format from protein data bank archive (https://www.rcsb.org/) and human antioxidant enzyme 1HD219 was used for investigate the antioxidant activity of EO from black pepper by docking method. The water molecules, heteroatoms, and inhibitors removed and hydrogen atoms added to the amino acid residues through PyMol software (ver. 2.4). Swiss-PdbViewer employed for minimize energy of the target proteins. Structure optimization done by Gaussian 09 software and the optimized structure subjected for docking study against selected proteins20.
ADME analysis
Pharmacokinetics properties of major component of essential oil from black pepper determined by online web tool SwissADME21.
Result and Discussion
Characterization of essential oil by GCMS
The yield of obtained essential oil founded 24.67% (w/w). The presence of volatile compounds in the EO identified by GCMS analysis comparing their retention time and molecular weight with the reference compounds in NIST mass spectra library (https://www.nist.gov/nist-research-library) and GCMS chromatogram shown in Fig 1. Thirteen compounds identified by GCMS analysis and present in Table 1. The major compounds identified as d-Norandrostane(5α,14α) (14.874%), Valencene (13.297%), 1-H-3a-7-Methanoazulene octahydro-1,9,9-trimethyl-4-methylene- (11.591%), (-)-spathulenol (8.193%), Aromandendrene (8.398%), and Napthalene,decahydro-4a-methyl-1-methylene-7-(1-methylethylidene) (7.794%) and shown in Fig 2. Other minor compounds identified as Bicyclo[7.2.0]undec-4-ene-4,11,11-trimethyl-8-methylene- (6.594%), Bicyclo[3.1.1]heptan-3-ol,6,6-dimethyl-2-methylene- (6.594%), 8-methylene-D-Limonene (5.159%), Linalool isobutyrate (3.728%), Alpha-Terpinol (3.692%), Azulene,1,2,3,3a,4,5,6,7-octahydro-1,4-dimethyl-7-(1-methylethenyl)- (2.890%), Caryophyllene oxide (2.578%), Ledene oxide-(II) (1.094%) and Disipro[2.1.2.4]undecane (1.458%).
 |
Figure 1: GCMS chromatogram of EO of black pepper. Click here to View figure |
 |
Figure 2: Chemical structure of major constituents in EO extract from black pepper. |
Table 1: Chemical constituents of EO extract of black pepper
|
Comp. |
Rt |
m/z |
(%) Area |
Similarity |
|
Disipro[2.1.2.4]undecane |
12.244 |
93.0 |
1.458 |
83 |
|
8-methylene-D-Limonene |
12.643 |
68.0 |
5.159 |
98 |
|
Linalool isobutyrate |
14.311 |
93.0 |
3.728 |
98 |
|
Alpha-Terpinol |
16.265 |
132.0 |
3.692 |
98 |
|
Bicyclo[3.1.1]heptan-3-ol, 6,6-dimethyl-2-methylene- |
16.557 |
91.0 |
5.168 |
95 |
|
Valencene |
18.075 |
136.0 |
13.297 |
84 |
|
Napthalene, 1,2,4a,5,6,8a-hexahydro-4,7-dimethyl-1-(1-methylethyl)- |
18.273 |
161.0 |
3.312 |
94 |
|
1-H-3a-7-Methanoazulene octahydro-1,9,9-trimethyl-4-methylene- |
18.821 |
161.0 |
11.591 |
98 |
|
Napthalene, decahydro-4a-methyl-1-methylene-7-(1-methylethylidene) |
19.062 |
147.0 |
7.794 |
98 |
|
Bicyclo[7.2.0]undec-4-ene-4,11,11-trimethyl-8-methylene- |
19.448 |
69.0 |
6.594 |
97 |
|
d-Norandrostane(5.alpha.,14.alpha) |
19.787 |
175.0 |
14.874 |
81 |
|
Aromandendrene |
20.299 |
107.0 |
8.398 |
97 |
|
Azulene, 1,2,3,3a,4,5,6,7-octahydro-1,4-dimethyl-7-(1-methylethenyl)- |
20.862 |
131.0 |
2.890 |
60 |
|
Caryophyllene oxide |
22.552 |
69.0 |
2.758 |
92 |
|
(-)-spathulenol |
23.004 |
131.0 |
8.193 |
96 |
|
Ledene oxide-(II) |
23.654 |
91.0 |
1.094 |
95 |
Antimicrobial activity test
In vitro antimicrobial activity test of EO determined with two numbers of Gram-positive bacteria, eight numbers of Gram-negative bacteria and six numbers of fungal strains using disc diffusion method. The results tabulate in Table 2 and Table 3 that obtained from this study. The extracted black pepper essential oil displayed moderate activity against used bacteria, compared to Ceftriaxone as standard (Table 2). The EO showed the highest activity with a zone of inhibition (20.7±0.6 mm) against Shigella boydii compared with standard Ceftriaxone (24.7±0.6 mm). The EO also showed good antifungal activity against all of the fungal strains with one exception Neurospora crassa (Table 3). The EO exhibited greater zone of inhibition with 13.7±1.5 mm against Trichodermal harzianum with standard Amphotericin B (10.7±1.5 mm).
Table 2: Antibacterial activity of EO extract of black pepper and Ceftriaxone.
|
Bacteria |
Strains |
Inhibition Zone (mm) |
|
|
EO |
Ceftriaxone |
||
|
Gram (+)ve bacteria |
Staphylococcus aureus |
16.0±1.0 |
40.3±0.6 |
|
Bacillus magneterium |
16.7±1.5 |
50.0±1.0 |
|
|
Gram (-)ve bacteria |
Salminella paratyphi |
19.0±1.0 |
49.0±1.0 |
|
Enterotoxigenic Escherica coli |
15.0±2.0 |
43.3±1.5 |
|
|
Salminella typhi |
18.0±1.0 |
42.7±1.5 |
|
|
Shigella flexneri |
14.3±2.5 |
37.7±1.5 |
|
|
Shigella sonnei |
16.3±0.6 |
39.7±0.6 |
|
|
Shigella boydii |
20.7±0.6 |
24.7±0.6 |
|
|
Escherica coli |
19.3±0.6 |
37.3±1.2 |
|
|
Shigella dysentary |
22.7±0.6 |
45.7±1.5 |
|
Table 3: Antifungal activity of EO extracts of black pepper and Amphotericin B.
|
Strains |
Inhibition Zone (mm) |
|
|
EO |
Amphotericin B |
|
|
Aspergillus niger |
11.0 ±1.0 |
15.0±1.0 |
|
Panysalium notatum |
7.3±1.2 |
15.3±0.6 |
|
Candida albican |
15.0±1.0 |
8.3±0.6 |
|
Aspergillus flavus |
10.3±2.5 |
16.0±1.0 |
|
Neurospora crassa |
– |
17.0±1.0 |
|
Trichodermal harzianum |
13.7±1.5 |
10.7±1.5 |
Antioxidant activity by DPPH assay
Antioxidant efficacy of EO determine by DPPH free radical scavenging method using a standard (ascorbic acid). IC50 values denote the desire concentration of sample, which inhibits 50% DPPH radicals. The concentrations and inhibitions of EO and ascorbic acid present in Fig 3. The IC50 values calculated from concentration against inhibition curves and EO shown good activity (IC50 35.83±2.92 µg/mL) compare with ascorbic acid (IC50 27.34±1.86 µg/mL) as standard (Table 4).
 |
Figure 3: Inhibition (%) DPPH radicals of EO extract of black pepper and ascorbic acid. |
Table 4: Antioxidant efficacy of EO extract of black pepper and Ascorbic acid.
|
Compounds |
IC50 (µg/mL) |
|
Essential Oil |
35.83±2.92 |
|
Ascorbic acid |
27.34±1.86 |
Molecular docking
Molecular docking performed with software Gaussian 09, PyRx 0.8, and Pymol for explore the interaction of target compound of EO with selected proteins. Protein structure optimization were done by DFT technique on B3LYP/6-31+G (d,p) basis set up. Compound 1 occupy highest percentage area (14.874%) in GCMS chromatogram (Table 1). Molecular docking study of compound 1 perform against selected PDB protein 1JIJ, 1KZN and 5JBO that are crystals of S. aureus, E. Coli and T. harzianum, respectively. Binding affinity of highest negative value chose for final appearance. The predicted interaction profiles of compound and target proteins illustrated in Fig 4. Compound 1 showed binding affinity -7.7 Kcal/mol, when docked against 1JIJ protein. It displayed Pi-sigma interactions with PHE306 at a distant of 3.50Å. In addition, several alkyl and Pi-alkyl interactions were involved with LYS305, PHE273, PHE 306, and PHE306 at a distance of 4.42 Å, 5.24 Å, 4.81 Å, and 4.98 Å, respectively. The binding affinity -6.2 Kcal/mol obtained when the compound 1 docked against 1KZN protein. It exhibited alkyl interactions with VAL120, LEU132, ILE78, LEU132, and LEU154 at a distance of 5.23 Å, 4.69 Å, 3.81 Å, 4.10 Å, and 4.10 Å, respectively. It also exhibited Pi-alkyl interaction with TYR145 at a distant of 5.13 Å. The highest binding affinity -9.0 Kcal/mol obtained when compound 1 docked against 5JBO protein. It showed several Pi-alkyl interactions with TRP357 at various distances. On the other hand, compound 1 showed binding affinity -6.9 Kcal/mol when docked with human peroxiredoxin 5 (antioxidant enzyme) 1HD2 protein (Fig 5) shown alkyl interactions with ARG86, ALA90, LEU96 at a distance of 4.73 Å, 4.12 Å, and 4.19 Å, respectively.
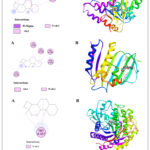 |
Figure 4: Molecular docking studies of compound 1 against 1JIJ, 1KZN, and 5JBO protein receptor. (A) 2D Interaction sketches. (B) 3D Docking prediction. |
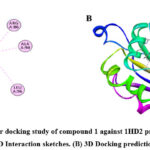 |
Figure 5: Molecular docking study of compound 1 against 1HD2 protein receptor. (A) 2D Interaction sketches. (B) 3D Docking prediction. Click here to View figure |
ADME analysis
To develop an effective drug in early preclinical trial is significant to know ADME (absorption, distribution, metabolism and excretion) including their pharmacokinetic drug like properties. Selected major six compounds (1–6) showed good ADME properties. The bioavailability score all of the compounds (>0.50) confirmed more drug-likeness properties. Most of the compounds showed low GI adsorption and BBB permeation except compound 4 and 5. Noticeably, there is no P-glycoprotein (P-gp) substrate to justify high GI adsorption. Compounds 4 and 5 can easily pass blood brain barrier (BBB), and can bind to specific receptors (Table 4). Any component showed better effectiveness with insignificant toxicity when they exhibited interactions with at least two isoenzymes of the cytochrome P (CYP) family. The compounds (1–6) with high affinities (low Kp) [(-5.44) to (-2.78)] describes higher strength of drug binding with receptor. The outcome of this study revealed that all selected compounds be strongly support for oral bioavailability. The radar chart of bioavailability placed inside the color zone with a small polygon closer to the centre represents pharmacokinetics, physicochemical and drug-likeness properties22. The radar plot chart of compounds (1–6) (Fig 6) denoted that all of the components are entirely inside the pink area, which indicates their good drug-likeness properties23.
Table 5: In silico ADME profile for major constituents of EO of black pepper.
|
Entry |
Compounds |
|||||
|
1 |
2 |
3 |
4 |
5 |
6 |
|
|
Lipiniski |
Yes |
Yes |
Yes |
Yes |
Yes |
Yes |
|
TPSA (Å2) |
0.00 |
0.00 |
0.00 |
0.00 |
20.23 |
0.00 |
|
Log(Po/w) |
5.45 |
4.41 |
4.61 |
4.34 |
3.26 |
4.06 |
|
Bioavailability score |
0.55 |
0.55 |
0.55 |
0.55 |
0.55 |
0.55 |
|
GI adsorption |
Low |
Low |
Low |
Low |
High |
Low |
|
BBB permeate |
No |
No |
No |
Yes |
Yes |
No |
|
P-gp substrate |
No |
No |
No |
No |
No |
No |
|
CYP1A2 inhibitor |
Yes |
No |
Yes |
Yes |
No |
No |
|
CYPC19 inhibitor |
Yes |
Yes |
Yes |
Yes |
Yes |
Yes |
|
CYP2C9 |
Yes |
Yes |
Yes |
Yes |
No |
Yes |
|
CYPD26 |
No |
No |
No |
No |
No |
No |
|
CYP134 |
No |
No |
No |
No |
No |
No |
|
log Kp (cm/s) |
-2.78 |
-3.83 |
-4.15 |
-4.20 |
-5.44 |
-4.42 |
 |
Figure 6: Radar plot chart of major constituents of EO of black pepper. Click here to View figure |
Conclusion
n-Hexane extract of black pepper essential oil showed moderate antibacterial activities and good antifungal and antioxidant activities that compare to their standard. The experimental results supported by in silico prediction and molecular docking studies. This n-hexane extract black pepper essential oil could be further screen as a potential source of biologically important drug candidates.
Acknowledgement
The authors acknowledge Chittagong University of Engineering & Technology, Chattogram-4349, Bangladesh for providing laboratory facilites.
Conflict of Interest
The authors declare that there is no potential conflict of interest.
References
- Hassan W, Kazmi, S.N.Z.; Noreen, H.; Riaz, A.; Zaman, B, J. Nutr. Disord. Ther., 2016, 6(2), 109. https://dx.doi.org/10.4172/2161-0509.1000190.
CrossRef - Sharma, R.N.; Xavier, F.P.; Vasu, K.K.; Chaturvedi, S.C.; Pancholi, S.S. J, Enzyme Inhib. Med. Chem., 2009, 24(3), 890–897. https://doi.org/ 10.1080/14756360802519558.
CrossRef - Tepe, B.; Dferera, D.; Solmen, M.; Polissou, M. and Sokmen, A, J. Agric. Food Chem., 2004, 52(5), 1132–1137. https://doi.org/10.1021/jf035094l.
CrossRef - Kordali, S.; Kotan, R.; Mavi, A.; Cakir, A.; Ala, A.; Yildirim, A, J. Agric. Food Chem., 2005, 53(24), 9452–9458. https://doi.org/10.1021/jf0516538.
CrossRef - Burt, S, Int. J. Food Microbiol., 2004, 94(3), 223–253. https://doi.org/10.1016/j.ijfoodmicro.2004.03.022.
CrossRef - Sylvestre, M.; Pichette, A.; Longtin, A.; Nagau, F.; Legault, J, J. Ethnopharmacol., 2006, 103(1), 99–102. https://doi.org/10.1016/j.jep.2005.07.011.
CrossRef - Mishra, A.K.; Mishra, A.; Kehri, H.K.; Sharma, B.; Pandey, A.K, Ann. Clin. Microbiol. Antimicrob., 2009, 8, 9. https://doi.org/10.1186/1476-0711-8-9.
CrossRef - Da Costa, A.C.; Dos Santos, B.H.C.; Filho, L.S.; Lima, E.D.O, Rev. Bras. Farmacogn., 2009, 19(1b), 236–241. https://doi.org/10.1590/S0102-695X2009000200010.
CrossRef - Jeena, K.; Liju, V.B.; Umadevi, N.P.; Kuttan, R, J. Essent. Oil-Bearing Plants., 2014, 17(1), 1–12. https://doi.org/10.1080/ 0972060X.2013.831562.
CrossRef - Ahmad, N.; Fazal, H.; Abbasi, B.H.; Farooq, S.; Ali, M.; Khan, M.A, Asian Pac. J. Trop. Biomed., 2012, 2(3), S1945–S1953. https://doi.org/10.1016/ S2221-1691(12)60524-3
CrossRef - Dinh, P.N.; Cam, H.D.T.; Quoc, T.P, IOP Conf. Ser.: Mater. Sci. Eng., 2020, 991, 012050. https://iopscience.iop.org/article/10.1088/1757-899X/991/1/012050/pdf
CrossRef - Rahman, M.M.; Alam, M. A.; Naher, S.; Jahan, A.; Ullah, A.K.M.A.; Khan, M.S.H, Int. J. Homeopath. Nat. Med., 2017, 3(2), 15-20.
http://dx.doi.org/ 10.11648/j.ijhnm.20170302.11
CrossRef - Rahman, M.; Khatun, A.; Islam, M.M.; Akter, M.N.; Chowdhury, S.; Khan, M.A.; Shahid, I.; Rahman, A, Int. J. Green Pharm., 2013, 7(3), 236–243. https://doi.org/10.4103/0973-8258.120242
CrossRef - Balouiri, M.; Sadiki, M.; Ibnsouda, S.K, J. Pharm. Anal., 2016, 6(2), 71–79. https://doi.org/10.1016/j.jpha.2015.11.005
CrossRef - Shah, M.S.; Rahman, M.M.; Islam, M.D.; Al-Macktuf, A.; Ahmed, J.U.; Nishino, H.; Haque, M.A, J. Mol. Struct., 2022, 1248, Article 131465. https://doi.org/10.1016/j.molstruc.2021.131465
CrossRef - Qiu, X.; Janson, C.A.; Smith, W.W.; Green, S.M.; McDevitt, P.; Johanson, K.; Carter, P.; Hibbs, M.; Lewis, C.; Chalker, A, Protein Sci., 2001, 10(10), 2008–2016. https://doi.org/10.1110/ps.18001
CrossRef - Wachino, J.; Jin, W.; Kimura, K.; Kurosaki, H.; Sato, A.; Arakawa, Y, mBio, 2020, 11(2), e03144-19, 1–17. https://doi.org/10.1128/mBio.03144-19
CrossRef - Florindo, R.N.; Souza, V.P.; Mutti, H.S.; Camilo, C.; Manzine, L.R.; Marana, S.R.; Polikarpov, I.; Nascimento, A.S, N. Biotechnol., 2018, 40(B), 218–227. https://doi.org/10.1016/j.nbt.2017.08.012
CrossRef - Declercq, J.P.; Evrard, C.; Clippe, A.; Stricht, D. Vander; Bernard, A.; Knoops, B, J. Mol. Biol., 2001, 311(4), 751–759. https://eurekamag.com/research/010/399/010399829.php
CrossRef - Hossain, R.; Sultana, R.; Din Islam, M.; Zaman, S.; Choudhary, M.I, Nat. Prod. Res., 2022, Online 19 Jan 2022, page 9. https://doi.org/10.1080/14786419.2022.2027934
CrossRef - Islam, M.D.; Kumar, S.; Chowdhury, T.A.; Sarker, M.Z.; Nishino, H.; Haque, M.A.; Rahman, M.M, J. Bangladesh Acad. Sci., 2021, 45(1), 37–47. https://doi.org/10.3329/jbas.v45i1.54258
CrossRef - Alam, A.; jawaid, T.; Alam, P, J. Taibah Univ. Sci., 2021, 15(1), 757–768. https://doi.org/10.1080/16583655.2021.2002550
CrossRef - Abishad, P.; Niveditha, P.; Unni, V.; Vergis, J.; Kurkure, N. V.; Chaudhari, S.; Rawool, D. B.; Barbuddhe, S. B, Gut Pathog., 2021, 13(1), 46, page 11. https://doi.org/10.1186/s13099-021-00443-3
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.









