Analysis of Volatile Metabolites Released by Staphylococcus Aureus using Gas Chromatography-Mass Spectrometry and Determination of its Antifungal Activity
Hayfaa Hussein Jaddoa1, Imad Hadi Hameed*2 and Ghaidaa Jihadi Mohammed3
1Department of Biology, Babylon University, Iraq.
2College of Nursing, Babylon University, Iraq.
3College of Science, Al-Qadisiya University, Iraq.
Corresponding Author E-mail: imad_dna@yahoo.com
DOI : http://dx.doi.org/10.13005/ojc/320439
Article Received on :
Article Accepted on :
Article Published : 20 Jul 2016
Bacterial volatile organic compounds (VOCs) have been considered as sensitive and specific biomarkers for bacterial detection in human specimens and culture media. The possibility of using VOCs markers as one of the largest groups of bacterial metabolites would open a new frontier for developing more efficient techniques in the diagnosis of bacterial infections. The aims of this research were analysis of the bioactive chemical products and evaluation of antibacterial and antifungal activity. Bioactives (chemical compounds often referred to as secondary metabolites) were analyzed using gas chromatography-mass spectroscopy (GC-MS) techniques, then the in vitro antibacterial and antifungal activity of the methanolic extract was evaluated. Thirty five bioactive compounds were identified in the methanolic extract of Staphylococcus aureus. GC-MS analysis of Staphylococcus aureus revealed the existence of the: Hexanoic acid , 2-methyl, 12,15-Octadecadiynoic acid , methyl ester, 1-Aminononadecane ,N-trifluoroacetyl-, N-[3-[N-Aziridyl]propylidene]hexylamine, N-(2,5-Dicyano-3,4-dihydro-2H-pyrrol-2-yl)-acetamide, 3-Cyclohex-3-enyl-propionic acid, 1-Methyl-4-[nitromethyl]-4-piperidinol, 3-Azonia-5-hexyn-1-ol , N,N-dimethyl-O-acetyl-,bromide, 1-Hexadecanol -2-methyl-, 1-Propyl-3,6-diazahomoadamantan, 9-Borabicyclo[3.3.1]nonane , 9-mercapto-, Benzyl methyl ketone, L-Aspartic acid ,N-glycyl-, Aminoacetamide , N-methyl-N-[4-(1-pyrrolidinyl)-2-butynyl]-, Tertbutyloxyformamide , N-methyl-N-[4-(1-pyrrolidinyl)-2-butyn, 5,7-Dodecadiyn-1,12-diol, Deoxyspergualin, D-Streptamine , O-6-deoxy-α-D-glucopyranosyl-(1-4), dl-Citrulline, N-[3-Diethylaminopropyl]-4-oxo-1,2,3,4,5,6,7,8-octahydroqui, N-Propionyl-D-glucoseamine, Cystine, 3,4-Dihydrocoumarin ,6-fluoro-4,4-dimethyl-, 4-(2,5-Dihydro-3-methoxyphenyl)butylamine, 3-methoxy-2-(1-methylethyl)-5-(2-methylpropyl)pyrazine, Uric acid, Thiocyanic acid 4-methoxy-2,6-dimethyl-3-pyridyl ester, 12-Dimethylamino-10-oxododecanoic acid, Glycyl-D-asparagine, Actinomycin C2, 12-Octadecenoic acid , methyl ester, 6-Octadecenoic acid , methyl ester ,(z)-, 2,5-Piperazinedione , 3,6-bis(2-methylpropyl)-, l-Leucyl-d-leucine and Methyl 12-hydroxy-9-octadecenoate. Gramineae poaceae was very highly active (6.71±0.13) mm. The results of anti-fungal activity produced by Staphylococcus aureus showed that the volatile compounds were highly effective to suppress the growth of Aspergillus terreus. Staphylococcus aureus produce many important secondary metabolites with high biological activities. Based on the significance of employing bioactive compounds in pharmacy to produce drugs for the treatment of many diseases, the purification of compounds produced by Staphylococcus aureus can be useful.
KEYWORDS:Antifungal activity; Staphylococcus aureus; GC-MS; Secondary metabolites
Download this article as:| Copy the following to cite this article: Jaddoa H. H, Hameed I. H, Mohammed G. J. Analysis of Volatile Metabolites Released by Staphylococcus Aureus using Gas Chromatography-Mass Spectrometry and Determination of its Antifungal Activity. Orient J Chem 2016;32(4). |
| Copy the following to cite this URL: Jaddoa H. H, Hameed I. H, Mohammed G. J. Analysis of Volatile Metabolites Released by Staphylococcus Aureus using Gas Chromatography-Mass Spectrometry and Determination of its Antifungal Activity. Orient J Chem 2016;32(4). Available from: http://www.orientjchem.org/?p=19317 |
Introduction
Staphylococcus aureus is a Gram-positive spherical bacterium approximately 1 μm in diameter. On a rich medium, S. aureus forms medium size “golden” colonies. One of the important characteristics of S. aureus is its capability to secrete toxins that disrupt membranes of host cells. Cytolytic toxins form β-barrel pores in the cytoplasmic membranes and cause leakage of the cell’s content and lysis1-3. Its cells form grape-like clusters, since cell division takes place in more than one plane. It is often found as a commensal associated with skin, skin glands, and mucous membranes, particularly in the nose of healthy individuals4. S. aureus secrets several cytolytic toxins, among them alpha-hemolysin, beta-hemolysin, gamma-hemolysin, leukocidin, and Panton-Valentine leukocidin (PVL)5. It has been estimated that approx. 20–30% of the general population are S. aureus carriers6. Staphylococcus aureus can be found in the anterior nares of a great proportion of a healthy human population. S. aureus is one of the main causes of hospital- and community-acquired infections which can result in serious consequences7,8. S. aureus is also able to cause a variety of skin and soft tissue infections and debilitating or even fatal diseases, such as pneumonia, necrotising fasciitis and septicaemia. Nosocomial S. aureus infections affect the bloodstream, skin, soft tissues and lower respiratory tracts. It can also produce toxins which cause toxin-mediated conditions such as toxic shock syndrome or food intoxications. It is a challenge to define which genetic factors determine whether an encounter between a human and S. aureus results in asymptomatic carriage or in clinical disease9-11. Many bacterial pathogens could lead to life-threatening infections. Accurate and rapid diagnosis is essential for the successful management of these infectious diseases. Traditional bacterial identification methods are time-consuming, require specific techniques and expertise. Other limitations of these techniques such as unaffordability and unavailability of expensive microbiological equipment and delay in the transport of human specimens such as fecal samples in diarrhea patients to the appropriate laboratories remain as main causes of delay in proceeding suitable curative actions in some countries. Therefore, all mentioned reasons have led to unavoidable delay in diagnosis and even death of infected patients12. Chemical analysis of bacterial culture includes analysis of bacterial metabolites, bacterial cell wall compositions and fatty acids profiling, have been introduced as bacterial differentiation and detection methods13-15. Metabolomics is a fast developing ‘omics’ that analyzes final metabolites of the cells by means of high throughput analytical technologies such as gas chromatography-mass spectrometry and high performance liquid chromatography-mass spectrometry16,17. Recent advances in ionization technologies allow researchers to perform sensitive qualitative and quantitative analysis of high molecular weight compounds along with the conventional ability of low molecular weight compound analysis in biological experiments18-21. The objectives of this research were analysis of the bioactive chemical products and evaluation of antibacterial and antifungal activity.
Materials and Methods
Growth conditions and determination of metabolites
S. aureus strain was isolated from bronchitis patients and obtained from Maternity and children hospital. Subcultures were obtained on the Nutrient Agar for 48 hrs. at 22°C. The mixture was incubated at 4ºC for 10 min. and then shook for 10 min. at 130 rpm. Metabolites was separated from the liquid culture and evaporated to dryness with a rotary evaporator at 45ºC. The residue was dissolved in 1 ml methanol, filtered through a 0.2 μm syringe filter, and stored at 4ºC for 24 h before being used for GC-MS. The identification of the components was based on comparison of their mass spectra with those of NIST mass spectral library as well as on comparison of their retention indices either with those of authentic compounds or with literature values22-26.
The studied fungi, Aspergillus niger, Aspergillus terreus, Aspergillus flavus, Aspergillus fumigatus, Candida albicans, Saccharomyces cerevisiae, Penicillium expansum, and Trichoderma viride were isolated and maintained in potato dextrose agar slants. Spores were grown in a liquid culture of potato dextrose broth (PDB) and incubated at 25ºC in a shaker for 16 days at 130 rpm. The extraction was performed by adding 25 ml methanol to 100 ml liquid culture in an Erlenmeyer flask after the infiltration of the culture.
Collection and Preparation of Plant Material
In this research, the leaves were dried at room temperature for ten days and when properly dried the leaves were powdered using clean pestle and mortar, and the powdered plant was size reduced with a sieve. The fine powder was then packed in airtight container to avoid the effect of humidity and then stored at room temperature27-31.
Extraction and Identification of Alkaloids
The powdered leaves (2 g) were boiled in a water bath with 20 ml of 5% sulphuric acid in 50% ethanol. The mixture was cooled and filtered. A portion was reserved. Another portion of the filtrate was put in 100 ml of separating funnel and the solution was made alkaline by adding two drops of concentrated ammonia solution. Equal volume of chloroform was added and shaken gently to allow the layer to separate. The lower chloroform layer was run off into a second separating funnel32-33. The ammoniacal layer was reserved. The chloroform layer was extracted with two quantities each of 5 ml of dilute sulphuric acid. The various extracts were then used for the following test:
Wagner’s test: To the filtrate in tube III, 1 ml of wagner’s reagent was added drop by drop. Formation of a reddish-brown precipitate indicates the presence of alkaloids.
Dragendoff’s test: To the filtrate in test tube II, 1 ml of dragendoff’s reagent was added drop by drop. Formation of a reddish-brown precipitate indicates the presence of alkaloids.
Mayer’s test: To the filtrate in test tube I, 1 ml of mayer’s reagent was added drop by drop. Formation of a greenish coloured or cream precipitate indicates the presence of alkaloids.
Spectral analysis of bioactive chemical compounds using gas chromatography-mass spectrometry (GC/MS)
Analysis was conducted using GC-MS (Agilent 789 A) equipped with a DB-5MS column (30 m×0.25 mm i.d., 0.25 um film thickness, J&W Scientific, Folsom, CA). The oven temperature was programmed as for the previous analysis. Helium was used as the carrier gas at the rate of 1.0 mL/min. Effluent of the GC column was introduced directly into the source of the MS via a transfer line (250oC). Ionization voltage was 70 eV and ion source temperature was 230oC. Scan range was 41- 450 amu. The components were identified by comparing their retention times to those of authentic samples of WILEY MASS SPECTRAL DATA BASE Library34-38.
Determination of antibacterial activity
Five-millimeter diameter wells were cut from the agar using a sterile cork-borer, and 25 μl of the samples solutions (Piper nigrum, Zingiber officinale, Gramineae poaceae, Nerium olender, Ricinus communis, Datura stramonium, Linum usitatissimum, Anastatica hierochuntica, Cassia angustifolia, Euphorbia lathyrus, Rosmarinus oficinalis, Mentha viridis, Artemisia annua, Quercus infectoria, Citrullus colocynthis, Althaea rosea, Coriandrum sativum, Origanum vulgare, Urtica dioica, Equisetum arvense, Foeniculum vulgare, Nigella sativa, Ocimum basilicum, Punica granatum, Punica granatum and Cinnamomum zeylanicum) were delivered into the wells. The plates were incubated for 48 h at room temperature. Antimicrobial activity was evaluated by measuring the zone of inhibition against the test microorganisms. Methanol was used as solvent control. The tests were carried out in triplicate. The antifungal activity was evaluated by measuring the inhibition-zone diameter observed after 48 h of incubation39-49.
Data analysis
All the measurements were replicated three times for each assay and the results are presented as mean ± SD and mean ± SE. IBM SPSS 20 version statistical software package was used for statistical analysis of percentage inhibition and disease incidence and disease severity in each case.
Results and Discussion
Gas chromatography and mass spectroscopy analysis of compounds was carried out in methanolic extract of S. aureus, shown in Table 1. The GC-MS chromatogram of the thirty one peaks of the compounds detected was shown in Figure 1.
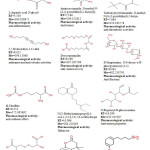 |
Table 1: Bioactive chemical compounds identified in methanolic extract of Staphylococcus aureus. Click here to View table |
 |
Figure 1: GC-MS chromatogram of methanolic extract of Staphylococcus aureus. chromatogram of methanolic extract of Staphylococcus aureus. Click here to View Figure |
Peaks were determined to be Hexanoic acid , 2-methyl, 12,15-Octadecadiynoic acid , methyl ester, 1-Aminononadecane ,N-trifluoroacetyl-, N-[3-[N-Aziridyl]propylidene]hexylamine, N-(2,5-Dicyano-3,4-dihydro-2H-pyrrol-2-yl)-acetamide, 3-Cyclohex-3-enyl-propionic acid, 1-Methyl-4-[nitromethyl]-4-piperidinol, 3-Azonia-5-hexyn-1-ol , N,N-dimethyl-O-acetyl-,bromide, 1-Hexadecanol -2-methyl-, 1-Propyl-3,6-diazahomoadamantan, 9-Borabicyclo[3.3.1]nonane , 9-mercapto-, Benzyl methyl ketone, L-Aspartic acid ,N-glycyl-, Aminoacetamide , N-methyl-N-[4-(1-pyrrolidinyl)-2-butynyl]-, Tertbutyloxyformamide , N-methyl-N-[4-(1-pyrrolidinyl)-2-butyn, 5,7-Dodecadiyn-1,12-diol, Deoxyspergualin, D-Streptamine , O-6-deoxy-α-D-glucopyranosyl-(1-4), dl-Citrulline, N-[3-Diethylaminopropyl]-4-oxo-1,2,3,4,5,6,7,8-octahydroqui, N-Propionyl-D-glucoseamine, Cystine, 3,4-Dihydrocoumarin ,6-fluoro-4,4-dimethyl-, 4-(2,5-Dihydro-3-methoxyphenyl)butylamine, 3-methoxy-2-(1-methylethyl)-5-(2-methylpropyl)pyrazine, Uric acid, Thiocyanic acid 4-methoxy-2,6-dimethyl-3-pyridyl ester, 12-Dimethylamino-10-oxododecanoic acid, Glycyl-D-asparagine, Actinomycin C2, 12-Octadecenoic acid , methyl ester, 6-Octadecenoic acid , methyl ester ,(z)-, 2,5-Piperazinedione , 3,6-bis(2-methylpropyl)-, l-Leucyl-d-leucine and Methyl 12-hydroxy-9-octadecenoate.
Antibacterial and antifungal activity
The results of anti-fungal activity produced by Staphylococcus aureus showed that the volatile compounds were highly effective to suppress the growth of Aspergillus terreus. Staphylococcus aureus produce many important secondary metabolites with high biological activities. Based on the significance of employing bioactive compounds in pharmacy to produce drugs for the treatment of many diseases, the purification of compounds produced by Staphylococcus aureus can be useful. Maximum zone formation against Aspergillus terreus (6.08±0.29) mm, Table 2. In agar well diffusion method the selected medicinal plants (Piper nigrum, Zingiber officinale, Gramineae poaceae, Nerium olender, Ricinus communis, Datura stramonium, Linum usitatissimum, Anastatica hierochuntica, Cassia angustifolia, Euphorbia lathyrus, Rosmarinus oficinalis, Mentha viridis, Artemisia annua, Quercus infectoria, Citrullus colocynthis, Althaea rosea, Coriandrum sativum, Origanum vulgare, Urtica dioica, Equisetum arvense, Foeniculum vulgare, Nigella sativa, Ocimum basilicum, Punica granatum, Punica granatum and Cinnamomum zeylanicum) were effective against Staphylococcus aureus, Table 3. Gramineae poaceae was very highly active (6.71±0.13) mm against Staphylococcus aureus. Staphylococcus aureus was found to be sensitive to all test medicinal plants and mostly comparable to the standard reference antifungal drug Amphotericin B and fluconazole to some extent. Recently, it was demonstrated that volatile organic compounds (VOCs) of bacteria such as terpenoids, phenylpropanoids and fatty acid derivatives can influence the growth of some fungi and, in general, the inter- and intra-organismic communication signals.
Table 2: Antifungal activity of Staphylococcus aureus metabolite products.
|
Fungi |
Antibiotics / Staphylococcus aureus metabolite products |
||||
|
|
Metabolite products of Staphylococcus aureus |
Amphotericin B |
Fluconazol |
Miconazole nitrate |
|
|
Aspergillus niger |
5.06±0.21 ª |
1.02±0.11 |
1.94±0.09 |
1.07±0.10 |
|
|
Aspergillus terreus |
6.08±0.29 |
2.06±0.16 |
1.02±0.16 |
2.01±0.12 |
|
|
Aspergillus flavus |
5.06±0.16 |
1.04±0.09 |
2.69±0.24 |
1.87±0.14 |
|
|
Aspergillus fumigatus |
5.67±0.20 |
0.97±0.10 |
3.01±0.14 |
1.01±0.13 |
|
|
Candida albicans |
5.01±0.11 |
1.48±0.12 |
1.79.±0.13 |
1.05±0.17 |
|
|
Saccharomyces cerevisiae |
3.03±0.17 |
1.02±0.12 |
1.03±0.10 |
2.13±0.12 |
|
|
Penicillium expansum |
2.68±0.14 |
2.03±0.20 |
3.90±0.15 |
0.86±0.11 |
|
|
Trichoderma viride |
3.35±0.20 |
2.06±0.11 |
1.37±0.12 |
1.32±0.12 |
|
|
Trichoderma horzianum |
3.02±0.19 |
0.39±0.01 |
2.05±0.13 |
1.97±0.16 |
|
ª The values ( average of triplicate) are diameter of zone of inhibition at 100 mg/mL methanolic extract and 30 μg/mL of (Amphotericin B; Fluconazol and Miconazole nitrate).
Table 3: Zone of inhibition (mm) of test different bioactive compounds and standard antibiotics of medicinal plants to Staphylococcus aureus.
| S. No. | Plant |
Zone of inhibition (mm) |
| 1. | Piper nigrum (Crude) |
5.35±0.19 |
| 2. | Zingiber officinale (Crude) |
4.22±0.20 |
| 3. | Gramineae poaceae (Crude) |
6.71±0.13 |
| 4. | Nerium olender (Alkaloids) |
3.36±0.20 |
| 5. | Ricinus communis (Alkaloids) |
1.86±0.19 |
| 6. | Datura stramonium(Alkaloids) |
2.52±0.20 |
| 7. | Linum usitatissimum (Crude) |
3.88±0.14 |
| 8. | Anastatica hierochuntica (Crude) |
4.17±0.18 |
| 9. | Linum usitatissimum (Crude) |
3.43±0.13 |
| 10. | Cassia angustifolia (Crude) |
5.51±0.25 |
| 11. | Euphorbia lathyrus (Crude) |
4.69±0.27 |
| 12. | Rosmarinus oficinalis (Crude) |
4.35±0.15 |
| 13. | Mentha viridis (Crude) |
3.29±0.14 |
| 14. | Artemisia annua (Crude) |
2.46±0.28 |
| 15. | Quercus infectoria (Crude) |
5.19±0.10 |
| 16. | Citrullus colocynthis (Crude) |
3.00±0.20 |
| 17. | Althaea rosea (Crude) |
4.70±0.15 |
| 18. | Coriandrum sativum (Crude) |
4.09±0.20 |
| 19. | Melia azedarach (Crude) |
2.76±0.24 |
| 20. | Origanum vulgare (Crude) |
5.15±0.26 |
| 21. | Urtica dioica (Crude) |
2.11±0.13 |
| 22. | Equisetum arvense (Crude) |
4.00±0.12 |
| 23. | Foeniculum vulgare (Crude) |
2.25±0.15 |
| 24. | Nigella sativa (Crude) |
3.27±0.24 |
| 25. | Ocimum basilicum (Crude) |
3.18±0.15 |
| 26. | Punica granatum (Crude) |
5.36±0.20 |
| 27. | Cinnamomum zeylanicum (Crude) |
3.01±0.18 |
| 28. | Streptomycin |
2.17±0.10 |
| 29. | Rifambin |
1.04±0.11 |
| 30. | Control |
00.00 |
Conclusion
Thirty five bioactive chemical constituents have been identified from methanolic extract of the Staphylococcus aureus by gas chromatogram mass spectrometry (GC-MS). In vitro antifungal and antibacterial evaluation of secondary metabolite products of Staphylococcus aureus forms a primary platform for further phytochemical and pharmacological investigation for the development of new potential antimicrobial compounds.
Acknowlgements
I would like to express my deep appreciation to head of department Prof. Dr. Ali Al-Marzooqi and Dean of the Science College Prof. Dr. Inas Al-Rubae for their support and encouragement.
References
- Foster, T.J. Nat Rev Microbiol. 2005, 3, 948–958.
CrossRef - Mohammed, G.J.; Kadhim, M.J.; Hussein, H.M. International Journal of Pharmacognosy and Phytochemical Research. 2016, 8(6), 889-905.
- Filipiak, W.; Sponring, A.; Baur, M.M.; Filipiak, A.; Ager, C. BMC Microbiol. 2012, 12, 113.
CrossRef - Buszewski, B.; Ulanowska, A.; Ligor, T.; Jackowski, M.; Klodzinska, E. Journal of Chromatography B: Biomedical Sciences and Applications. 2008, 868, 88-94.
CrossRef - Kaneko, J.; Kamio, Y. Biosci Biotechnol Biochem. 2004, 68, 981–1003.
CrossRef - Kunze, N.; Göpel, J.; Kuhns, M.; Jünger, M.; Quintel, M. Applied microbiology and biotechnology. 2013, 97, 3665-3676.
CrossRef - Schulz, S.; Dickschat, J.S. Nat Prod Rep. 2007, 24, 814-842.
CrossRef - Zhu, J.; Bean, H.D.; Kuo, Y.M.; Hill, J.E. J Clin Microbiol. 2010, 48, 4426-4431.
CrossRef - Becker, K.; Friedrich, A.W.; Lubritz, G.; Weilert, M.; Peters, G.; Von Eiff , C. J Clin Microbiol. 2003, 41, 1434–1439.
CrossRef - Fueyo, J.M., Mendoza, M.C., Martín, M.C. Microbes Infect , 2005, 7, 187–194.
CrossRef - Belkum, A.; Melles, D.C.; Nouwen, J., Leeuwen, W.B.; Wamel, W., Vos, M.C., Wertheim, H.F.; Verbrugh, H.A. Infect Genet Evol, 2009, 9, 32–47.
CrossRef - Probert, C.; Jones, P.; Ratcliffe, N. GUT, 2004, 53, 58-61.
CrossRef - Ehrhardt, C.J.; Chu, V.; Brown, T.; Simmons, T.L.; Swan, B.K. Applied and Environmental Microbiology, 2010, 76, 1902-1912.
CrossRef - Li, Y., Wu, S., Wang, L.; Li, Y.; Shi, F. J Sci Food Agric. 2010, 90, 1380-1383.
CrossRef - Pagans, E.; Font, X.; Sánchez, A. Journal of hazardous materials. 2006, 131, 179-186.
CrossRef - Wang, D.; Ding, X.; Rather, P.N. Journal of Bacteriology. 2001, 183, 4210-4216.
CrossRef - Savelev, S.U.; Perry, J.D.; Bourke, S.J.; Jary, H.; Taylor, R. Lett Appl Microbiol. 2011, 52, 610-613.
CrossRef - Glish, G.L.; Vachet, R.W. Nature Reviews Drug Discovery. 2003, 2, 140-150.
CrossRef - Ulanowska, A.; Kowalkowski, T.; Hrynkiewicz, K.; Jackowski, M.; Buszewski, B. Biomed Chromatogr, 2011, 25, 391-397.
CrossRef - Garner, C.E.; Smith, S.; Elviss, N.C.; Humphrey, T.J.; White, P. Biomarkers. 2008, 13, 413-421.
CrossRef - Davies, J.C. Paediatr Respir Rev. 2002, 3, 128-134.
CrossRef - Liu, G.Y.; Essex, A.; Buchanan, J.T.; Datta, V.; Hoffman, H.M.; Bastian, J.F. J Exp Med 2005, 202, 209–215.
CrossRef - Altameme, H.J.; Hameed, I.H.; Abu-Serag, N.A. Malaysian Applied Biology, 2015b, 44(4), 47–58.
- Mohammed, G.J.; Al-Jassani, M.J.; Hameed, I.H. International Journal of Pharmacognosy and Phytochemical Research, 2016, 8(3), 480-494.
- Hussein, H.J.; Hadi, M.Y.; Hameed, I.H. Journal of Pharmacognosy and Phytotherapy, 2016, 8(3), 60-89.
CrossRef - Shareef, H.K.; Muhammed, H.J.; Hussein, H.M.; Hameed, I.H. Orient J Chem, 2016, 32(2), 20-40.
- Hamza, L.F.; Kamal, S.A.; Hameed, I.H. Journal of Pharmacognosy and Phytotherapy, 2015, 7(9), 194-220.
CrossRef - Al-Jassaci, M.J.; Mohammed, G.J.; Hameed, I.H. International Journal of Pharmaceutical and Clinical Research, 2016, 8(5), 304-315.10.
- Hameed, I.H.; Ibraheam, I.A.; Kadhim, H.J. Journal of Pharmacognosy and Phytotherapy, 2015c, 7(6), 90-106.
CrossRef - Al-Marzoqi, A.H.; Hadi, M.Y.; Hameed, I.H. Journal of Pharmacognosy and Phytotherapy, 2016, 8(2), 25-48.
CrossRef - Hadi, M.Y.; Mohammed, G.J.; Hameed, I.H. Journal of Pharmacognosy and Phytotherapy, 2016, 8(2), 8-24.
CrossRef - Kadhim, M.J.; Mohammed, G.J.; Hameed, I.H. Orient J Chem. 2016; 32(2), 10-30.
- Jasim, H.; Hussein, A.O.; Hameed, I.H.; Kareem, M.A. Journal of Pharmacognosy and Phytotherapy, 2015, 7(4), 57-72.
- Altameme, H.J.; Hameed, I.H.; Idan, S.A.; Hadi, M.Y. Journal of Pharmacognosy and Phytotherapy. 2015c, 7(9), 222-237.
- Hussein, A.O.; Mohammed, G.J.; Hadi, M.Y.; Hameed, I.H. Journal of Pharmacognosy and Phytotherapy, 2016, 8(3), 49-59.
CrossRef - Hameed, I.H.; Altameme, H.J.; Idan, S.A. Research Journal of Pharmaceutical, Biological and Chemical Sciences. 2016, 7(2), 1843- 1868.
- Ratjen, F. Current opinion in pulmonary medicine, 2006, 12, 428-432.
CrossRef - Shestivska, V., Španel, P.; Dryahina, K.; Sovová, K.; Smith, D. Journal of Applied Microbiology. 2012, 113, 701-713.
CrossRef - Zechman, J.M.; Aldinger, S.; Labows, J.N. Journal of Chromatography B: Biomedical Sciences and Applications, 1986, 377, 49-57.
CrossRef - Hameed, I.H.; Hussein, H.J.; Kareem, M.A.; Hamad, N.S. Journal of Pharmacognosy and Phytotherapy, 2015b, 7(7), 107-125.
CrossRef - Sosa, A.A.; Bagi, S.H.; Hameed, I.H. International Journal of Pharmacognosy and Phytochemical Research, 2016, 8(5), 109-126 ().
- Hussein, H.M.; Hameed, I.H.; Ibraheem, O.A. International Journal of Pharmacognosy and Phytochemical Research, 2016, 8(3), 369-385.
- Mohammed, G.J.; Omran, A.M.; Hussein, H.M. International Journal of Pharmacognosy and Phytochemical Research. 2016, 8(6), 977-996.
- Kadhim, M.J.; Sosa, A.A.; Hameed, I.H. International Journal of Pharmacognosy and Phytochemical Research, 2016, 8(6), 127-146.
- Altameme, H.J.; Hadi, M.Y.; Hameed, I.H. Journal of Pharmacognosy and Phytotherapy. 2015a, 7(10), 238-252.
CrossRef - Hameed, I.H.; Hamza, L.F.; Kamal, S.A. Journal of Pharmacognosy and Phytotherapy. 2015a, 7(8), 132-163.
CrossRef - Kaur, R.; Kaur, H. Orient J Chem. 2015, 31(1), 597-600.
- Chandrashekharaiah, K.S. OrientJ Chem. 2013, 29(3), 1061-1070.
- Kadhim, M.J.; Mohammed, G.J.; Hameed, I.H. Orient J Chem. 2016, 32(3).

This work is licensed under a Creative Commons Attribution 4.0 International License.









