Dereplication Study on Glaucium Aleppicum Boiss. in Jordan
S. A. Barakat*, Feras, Q. Al Ali and M. A. Al Hammouri
Jordan University of Science and Technology. 22110 Irbid, P.O.Box 3030, Jordan.
Corresponding Author E-mail: barakat@just.edu.jo
DOI : http://dx.doi.org/10.13005/ojc/320409
Article Received on :
Article Accepted on :
Article Published : 03 Aug 2016
The secondary metabolite constituents of Glaucium aleppicum Boiss. et Hausskn. ex Boiass. (Papaveraceae) were investigated using an in-house developed Ultra Performance Liquid Chromatography-Photodiode Array-High Resolution Tandem Mass Spectrometry (UPLC-PDA-HRMS-MS/MS) method. In this study the powerful resolution and short analysis time afforded by UPLC coupled to the outstanding mass accuracy of an Orbitrap mass spectrometer was used to develop a rapid and effective dereplication method to identify secondary metabolite constituents of G. aleppicum in crude extracts. In addition, HCD MS/MS and UV (photodiode array) spectra were used as an integral part of the strategy. This methodology was applied on a small amount of extract (~ 1 mg) of G. aleppicum to dereplicate twelve compounds. Five of which have been isolated previously from G. aleppicum: allocryptopine, protopine, corydine, isocorydine, and norchelidonine. Seven were new to the species, these include: dehydroglaucine, norisocorydine, chelerythrine, norsanguinarine, dihydrochelerythrine, corytuberine, and fumaflorine methyl ester. In addition to one more compound that was unidentified and thus considered new.
KEYWORDS:Norisocorydine; Norchelidonine; Dihydrochelerythrine; Corytuberine; Norsanguinarine; Fumaflorine methyl ester; Chelerythrine; Dehydroglaucine; Glucium aleppicum; LC-MS
Download this article as:| Copy the following to cite this article: Barakat S. A, Feras F, Al Ali Q, Al Hammouri M. A. Dereplication Study on Glaucium Aleppicum Boiss. in Jordan. Orient J Chem 2016;32(4). |
| Copy the following to cite this URL: Barakat S. A, Feras F, Al Ali Q, Al Hammouri M. A. Dereplication Study on Glaucium Aleppicum Boiss. in Jordan. Orient J Chem 2016;32(4). Available from: http://www.orientjchem.org/?p=19171 |
Introduction
Plants have been used as medicines for thousands of years. Historically, natural products have played an important role in treating and preventing diseases [1]. They have long been and will continue to be highly important as sources of medicinal agents and models for the design, synthesis, and semi-synthesis of novel substances for preventing and treating human diseases [2-5]. These medicines initially took the form of crude drugs such as teas, tinctures poultices, powders, and other herbal formulations [2-5]. Globally, traditional medicines depend mainly on herbal products. Drugs extracted from natural products mainly come from plants and microorganisms, to lesser extent from marines, invertebrates and vertebrates. The fast identification of known compounds present in a mixture is very important for rapid discovery of novel natural products, this is called dereplication. Dereplication speed up the discovery of lead compounds and saves time and resources [6-9]. Dereplication strategy normally combines chromatographic and spectroscopic methods and searching in database to discriminate between previously isolated compounds, and new compounds at the level of the crude extract [6-9]. Traditional natural drugs revelation programs utilize bioactivity-directed fractionation approaches to segregate, sanitize and describe bioactive lead compounds from crude extracts, wherein the bioassay results drive the purification processes. This process is often stodgy, costly, and time consuming; moreover, it may end up with isolation of a previously known bioactive compound; thus yielding no net earning [6,7,10,11, 12,13]. To speed up drug discovery timelines of new biologically active natural products. It is of great importance to discriminate between previously isolated, known compounds and new compounds, at the level of the crude extract [6,7,811,12]. Dereplication is an essential component of natural products studies, It is used to make sure that resources are expended on only the most promising samples and avoid the tedious isolation of known constituents, which are those most likely to yield bioactive and structurally unique compounds [13-17]. As a result, some molecular masses and the corresponding fragmentations do not correlate with any known compounds, thus revealing potentially novel natural products that could be investigated on a larger scale and could completely find application as new drugs [6]. Using molecular masses, their fragmentation patterns, the suitable database and LC-retention times, it is possible to dereplicate some alkaloid compounds by their structural elucidation when compared with known structures of natural origin [6]. Hyphenated techniques can be defined as the coupling of HPLC separation technologies with on-line spectroscopic/spectrometric detection technologies [18, 19]. The extraordinary improvements in hyphenated analytical methods over the past two decades have broadened their applications in the analyses of biomaterials significantly, especially natural products. Ultra Performance Liquid Chromatography (UPLC) have more advantages than HPLC because. It features by particle chemistry performance, system optimization, detector design and data processing and control which leads to increasing the overall interlaced attributes of speed, sensitivity, and resolution. This technique has the advantages of high quality, where it has small particles with very good packing and higher flow rates with superior resolution and sensitivity [20]. The Hashemite Kingdom of Jordan lies in the heart of the Middle East. It’s located in the junction between Asia, Africa and Europe, between latitudes 29º-33º N and longitudes 35º-39º E, 100 km from the southeastern coast of the Mediterranean Sea with four biogeographical areas: the Mediterranean, the Saharo-Arabian, the Tropical or Sudanian and the Irano-Turanean region [21, 22]. Jordan also covers a wide range of elevation from 400 m below the sea level near the Dead Sea to 1750 m above the sea level in the south mountains near to Jabal Rum. This place at the crossroad of the three continents, different biogeographical areas and different climate places supplies Jordan with diverse types of plant life reached to 700 genera with more than 2500 species, about 100 species considered as endemic species, 250 rare and 125 very rare species [23]. In this research, Glaucium aleppicum Boiss. et Hausskn. ex Boiss. was selected for LC-MS investigation. The specific objective were, analytically, identifying the alkaloids in Glaucium aleppicum Boiss. et Hausskn. ex Boiss. (Papaveraceae) and 2. Fully proofing the concept of fast dereplication strategy for Glaucium aleppicum Boiss. et Hausskn. ex Boiss. (Papaveraceae) species. The family Papaveraceae has 28th genera and four hundred fifty species and the genus Glaucium belongs to this family. The species of this family are annual or perennial herbs, rarely shrubs and trees, often its juice is colored, flourish mainly in the Northern hemisphere in warm areas and South America. Glaucium (Horned Poppy) is a genus of about 25 species of annuals and perennials, hairy, glaucous herbs with yellow juice [24].
Exeperimental
Ultra Performance Liquid Chromatography-Photodiode Array-High Resolution Tandem Mass Spectrometry (UPLC-PDA-HRMS-MS/MS):
HR-ESI-MS was performed on a Thermo LTQ Orbitrap XL mass spectrometer (ThermoFisher, San Jose, CA) equipped with an electrospray ionization source. Source conditions in the positive ionization mode were set at 275 °C for the capillary temperature, 4.5 kV for the source voltage, 20 V for capillary voltage, and 95 V for tube lens. Nitrogen was utilized for the sheath gas and set to 25 arb. Two scan events were carried out, full scan (100-500) and MS/MS of the most intense ion from the parent mass list utilizing HCD( Higher-energy collisional dissociation, an ion fragmentation technique) with a normalized collision energy ranging from 22-60. External instrument calibration was performed using an LTQ ESI positive ion calibration solution consisting of caffeine (20 µg/mL), MRFA (1 µg/mL) and Ultramark 1621 (0.001%) in an aqueous solution of CH3CN (50%), MeOH (25%) and acetic acid (1%). Thermo Scientific Xcalibur 2.1 software was used for instrument control and data analysis. UPLC was carried out on a Waters Acquity System [using a BEH C18 (2.1 × 50 mm, 1.7 µm) column (Waters Corp., Massachusetts, USA) equilibrated at 40 °C]. A mobile phase consisting of CH3CN:H2O (acidified with 0.1% formic acid) was used, starting with 5:95 with a hold for 1 min, then increasing linearly to 20% CH3CN within 3 min, then increasing linearly to 100% CH3CN within 4 min, holding for 1.5 min and then returning to the starting conditions within 0.1 min. Flow rate used was 0.3 mL/min with an injection volume of 1 µ. An acquity UPLC photodiode array detector was used to acquire PDA spectra, which were collected from 191-499 nm with 4 nm resolution.
Low Resolution Liquid Chromatography-Mass Spectrometry: LRSPCI-MS utilized an Applied Biosystem (API 3200) triple quardapole LC/MS coupled with APCI positive ionization mode and an Agilant® 1200 series HPLC equipped with a Hypersil ODS (150 mm × 4 mm; 5 µm) column (Thermo Electron, Auchtermuchty, UK). The mobile phase used was H2O (A) and MeOH (B) both acidified with 0.1% formic acid in the following three gradient combinations systems (I-III): Solvent
| System I | Solvent System II | ||||
| Time (min) | Water/ formic acid (%) | Methanol (%) | Time (min) | Ammonium formate/water (%) | Ammonium formate/Acetonitrile (%) |
| 00 | 80 | 20 | 0-3 | 100 | 0 |
| 10 | 80 | 20 | 3-6 | 10 | 90 |
| 30 | 20 | 80 | 6-10 | 10 | 90 |
| 45 | 20 | 80 | 10-12 | 100 | 0 |
| 50 | 80 | 20 | 12-15 | 100 | 0 |
| 60 | 80 | 20 |
The mass detector conditions were as follows: APCI positive ionization mode and ESI positive ionization mode, full scan mode from 50 to 800 m/z and 55 to 550 m/z, corona discharge voltage NC5 kv, APCI temperature 500 ºC, gas flow rate 5 mL/min. The mobile phase flow rate was 1 mL/min with an injection volume of 20 µL.
| Solvent System III | ||
| Time | Formic acid/ water (%) | Methanol (%) |
| 0 | 80 | 20 |
| 5 | 80 | 20 |
| 30 | 20 | 80 |
| 40 | 20 | 80 |
| 45 | 80 | 20 |
Instrumentation
High resolution MS data were acquired using a Thermo LTQ Orbitrap XL mass spectrometer (ThermoFisher, San Jose, CA) equipped with an electrospray ionization source. While low resolution MS data were collected using an Agilant® Applied Biosystem (API 3200) triple quadrupole LC/MS coupled with APCI positive ionization mode and an Agilent® 1200 series HPLC. UPLC was carried out on a Waters Acquity system [using a BEH C18 (2.1 × 50 mm, 1.7 µm) column (Waters Corp., Massachusetts, USA). Analytical HPLC columns used were an Eclips XDB C18 (150 mm × 4.6 mm; 5 µm), and a propyl PFP (50 mm × 2.1 mm; 5 µm). Samples were prepared using digital micropipettes, 1000-100 µL and 100-10 µL. Witeglaborterchik GmbH, Germany. Samples were evaporated using a rotary evaporator, RE 200, BIBBY, BIBBY STERILINE LTD, UK. Balances used were Mettler BasBal BB240 Balance, type BB 240, SNR M14187, FNR 215200 14204 and Sartorius analytic A120S. A Eurosonic sonicator 22 analis, Namur 081/225085, was used.
Reagents
Fisher Scientific Optima LC/MS acetonitrile and Water were used for UPLC-MS runs. Methanol and acetonitrile, both HPLC grade, were obtained from Fisher Scientific, U.K. Formic acid, analytical grade, was purchased from Scharlau Chemie S.A. Barcelona, Spain. Allocryptopine standard was obtained from Aldrich. Caffeine (20 µg/ml) was used for calibration.
Results and Discussion
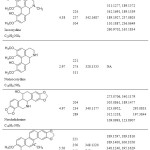 |
Table : Chemical structures, chemical formulas, retention times, UV absorption maxima, (+)-ESI HRMS, and (+)-ESI HCD MS/MS of the identified compounds from G. aleppicum. Click here to View Table |
 |
Figure 1: Spectrum Click here to View Figure |
Conclusion
A UPLC-PDA-HRMS-MS/MS methodology was developed and implemented successfully for the dereplication of alkaloids in crude extracts of G. aleppicums. The methodology has several advantages. First, the use of HRMS data imparts a high degree of confidence in the structure of the dereplicated hits. Second, the use of MS/MS HCD along with UV absorption maxima confirmed the identity of the dreplicated compounds. Third, the use of UPLC enables a rapid (10 min) chromatographic method, one of the fastest methods that have been utilized in dereplication methodologies of natural products. Finally, the use of the ACD/IntelliXtract, which extracts all chromatographic components in the LC-MS datasets, expedites the identification process. In particular, the capability of the ACD/IntelliXtract to deconvolute overlapping and co-eluting components makes it possible to dereplicate trace compounds and enables the application of the short (10 min) chromatographic run time.
In summary, we were successful in developing a short and efficient dereplication methodology that can differentiate between known and new compounds at the level of the crude extracts and hence expedite the discovery of new compounds. The method can be used for detection and identification of structurally related alkaloids from different natural products resources.
Aknowledgment
Many Thanks go to Deanship of Research, Jordan University of Science and Technology for their Financial support.
References
- Williamson E. M., Okpako D. T. and Evans. F. J. Selection, Preparation and pharmacological evaluation of plant material. New York: John wiley and sons; 1996: 1-7.
- Samuelsson G. Drugs of Natural Origin: a Textbook of Pharmacognosy, 5th Swedish Pharmaceutical Press, Stockholm; 2004.
- Balick, M.J., Cox, P.A. Plants, People, and Culture: the Science of Ethnobotany. Scientific American Library, New York, NY; 1997.
- Kinghorn, A.D. Pharmacognosy in the 21st century. Journal of Pharmacy and Pharmacology. 2001; 53 (2):135-148.
CrossRef - G. R. Hamilton, T. F. Baskett, Can. J. Anaesth. 2000. 47, 367
CrossRef - W.F. Smyth, T.J.P. Smyth, V.N. Ramachandran, F. O_Donnell, P. Brooks, Dereplication of phytochemicals in plants by LC-ESI-MS and ESI-MSn 2012, Trends Anal. Chem. 2012.
- Alali Feras Q, Gharaibeh Ahmad, Ghawanmeh Abdullah, Tawaha Khalid and Oberlies Nicholas,. Phytochem. Anal. (2008).
- Cordell Geoffrey A. ,. Pure Appl. Chem 1999 .,71, 1089 – 1094.
CrossRef - Nielsen Kristian F. , Nat. Prod. 2011,. 74, 2338 – 2348.
CrossRef - Mishra Bhuwan B. , Tiwari Vinod K. , Natural products: An evolving role in future drug discovery 2011, European Journal of Medicinal Chemistry 2011; 46: 4769-4807.
CrossRef - Ghisalberti E. Detection and isolation of bioactive natural products. In: Colegate S, Molyneux R, eds. Detection, isolation and structural determination: CRC Press, Inc. 1993:9-57.
- Kingston D. Natural produts as pharmaceuticals and sources for lead structures. In: Meyer P, ed. The practice of Medicinal Chemistry: Academic Press Limited. 1996:101-16.
- Cordell GA, Shin YG. Finding the needle in the haystack. The dereplication of natural product extracts. Pure Applied Chemistry. 1999; 71:1089-94.
CrossRef - Hostettmann K, Wolfender J, Terreaux C. Modern screening techniques for plant extracts. Pharmaceutical Biology. 2001; 39:18-32.
- Wolfender J, Queiroz EF, Hostettmann K. The importance of hyphenated techniques in discovery of new lead compounds from nature. Expert Opinion on Drug Discovery. 2006; 1:237-60.
CrossRef - Ackermann B, Regg B, Colombo L, Stella S, Coutant J. Rapid analysis of antibiotic-containing mixtures from fermentation broths by using liquid chromatography-Electrospray ionization-mass spectrometry and matrix-assisted laser desorption ionization-time-of-flight-mass spectrometry. J Am Soc Mass Spectrum. 1996; 7:1227-37.
CrossRef - Constant H, Beecher C. A method for the dereplication of natural product extracts using electrospray HPLC/MS. Natural Product Letters. 1995; 6:193-6.
CrossRef - Wilkins, C. L. Hyphenated techniques for the analysis of complex organic mixtures. Science 222, 1983:291-296.
CrossRef - Robert E. Ardrey. liquid chromatography – mass spectrometry: an introduction, 2003:1-276.
- Michael E. Swartz, Ultra Performance Liquid Chromatography (UPLC): An Introduction, chromatography line; 2005; 8 – 14.
- Al-Eisawi D. M. Field Guide to wild Flowers of Jordan and Neighboring Countries. Amman, Jordan: Jordan Press, Foundation Al-Rai; 1998:1-2.
- Dothan N. F. Palestina Flora. Jerusalim: The Israel Academy of Science and Humanities; 1996: 43.
- Anonymous. Conservation and Sustainable Use of Biological Diversity in Jordan, The Hashemite Kingdom of Jordan, Amman; 2001.
- Fedde F., Papaveraceae-Hypecoideae et Papaveraceae- Papaveroideae, in: Pflznr. 1909; 40: 1-430.

This work is licensed under a Creative Commons Attribution 4.0 International License.









